10.1.1 使用python完成第一个遗传算法
举例如下:![]()
求函数的在x=[-3,3],y=[-3,3]上的最大值:
【思路】
1. 把DNA设为:48个bit的数组。 奇数为x, 偶数为y;就是说24个位组成x,24个位组成y。
如何得到x,y: 24个奇数位转换成数值比如b, 则 x = b/(2^24-1)* (3 - (-3)) -3
2.交叉因子:
就是父的前一部分+母DNA的后一部分
3. 变异因子:
就是某个位取反。
【代码】
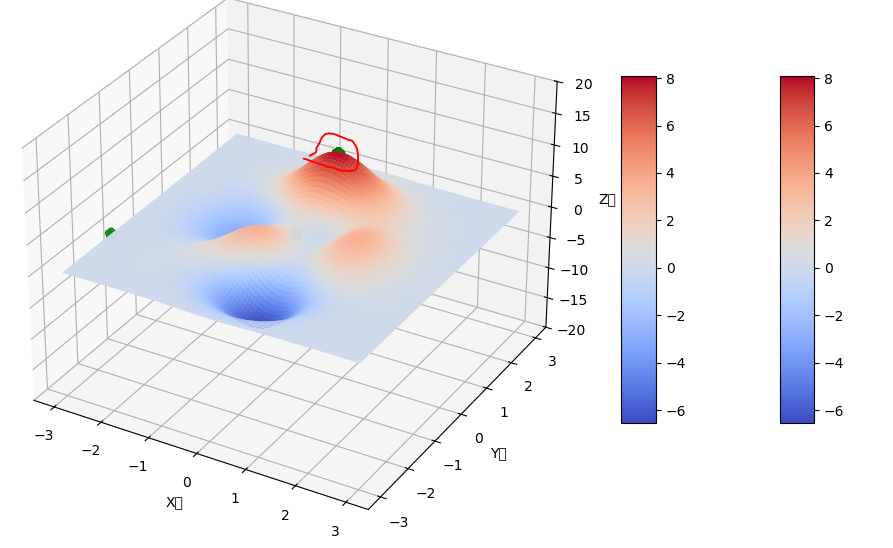
代码如下,使用200条DNA,迭代100次,得到结果 (x,y)=(0,1.582), z=8.1. 结果比较稳定。
import numpy as np
import matplotlib.pyplot as plt
from jmespath.ast import projection
from matplotlib import cm
from mpl_toolkits.mplot3d import Axes3DDNA_SIZE=24
POP_SIZE=200
N_GENERATIONS=100
CROSSOVER_RATE=0.8
MUTATION_RATE=0.005X_BOUND =[-3,3]
Y_BOUND =[-3,3]
def F(x,y):# 2(3-x)^2 e( -x^2 - (y+1)^2) - 4 e(-(x+2)^2-y^2) - 6 e(-(x-2)^2-y(y-2)^2)#return 2*(3-x)**2* np.exp(-x**2-(y+1)**2)-4*np.exp(-(x+2)**2-y**2)-6*np.exp(-(x-2)**2-(y-2)**2)return 3*(1-x)**2*np.exp(-(x**2)-(y+1)**2)-10*(x/5-x**3-y**5)*np.exp(-x**2-y**2)-1/2*np.exp(-(x+1)**2-y**2)#return 10-x**2-y**2
def plot_3d(ax):x = np.linspace(*X_BOUND, 100)y = np.linspace(*Y_BOUND, 100)X, Y = np.meshgrid(x, y)Z = F(X, Y)surf = ax.plot_surface(X,Y,Z,rstride=1,cstride=1,cmap = cm.coolwarm)# 添加颜色条fig.colorbar(surf, ax=ax, shrink=0.5, aspect=10)# 设置坐标轴标签ax.set_zlim((-20,20))ax.set_xlabel('X轴')ax.set_ylabel('Y轴')ax.set_zlabel('Z轴')# 设置标题ax.set_title(':z=f(x,y)')plt.pause(3)plt.show()def translateDNA(pop):x_pop = pop[:,1::2] #奇数列表示xy_pop = pop[:,::2] #偶数列表示yx = x_pop.dot( 2** np.arange(DNA_SIZE) [::-1] )/float(2**DNA_SIZE-1)*(X_BOUND[1]-X_BOUND[0])+ X_BOUND[0]y = y_pop.dot( 2** np.arange(DNA_SIZE) [::-1] )/float(2**DNA_SIZE-1)*(Y_BOUND[1]-Y_BOUND[0])+ Y_BOUND[0]return x,ydef crossover_and_mutaion(pop,Crossover_rate=CROSSOVER_RATE):halfpop = int(POP_SIZE/2)fitness = get_fitness(pop)indices = np.argpartition(fitness, -halfpop)[-halfpop:] #得到前50%最好的样本进行保留new_pop=[]#保留原来比较好的50%.for pepole in pop[indices]:new_pop.append(pepole)#其他的进行交叉以及变异for father in pop:child = fatherif(np.random.rand()<Crossover_rate):mother = pop[np.random.randint(POP_SIZE)]cross_point=np.random.randint(0,DNA_SIZE*2)child[cross_point:]=mother[cross_point:]mutation(child) #变异new_pop.append(child)return new_pop#编译
def mutation(child,mutation_rate=MUTATION_RATE):if np.random.rand()<mutation_rate:mutation_point = np.random.randint(0,DNA_SIZE*2)child[mutation_point]= child[mutation_point]^1#选择
def select(pop,fitness):idx = np.random.choice(np.arange(fitness.size),size=POP_SIZE,replace=True,p=(fitness)/fitness.sum())return pop[idx]def get_fitness(pop):x,y=translateDNA(pop)pred=F(x,y)return (pred- np.min(pred))+1e-3def print_info(pop):fitness = get_fitness(pop)max_fitness_index= np.argmax(fitness)x,y=translateDNA(pop)print("最优基因:",pop[max_fitness_index])print("(x,y):",(x[max_fitness_index],y[max_fitness_index]))if __name__ == '__main__':# main函数,程序入口# 创建数据fig = plt.figure(figsize=(12,9))ax=fig.add_subplot(111,projection='3d')plt.ion()plot_3d(ax)population = np.random.randint(2,size=(POP_SIZE,DNA_SIZE*2))for i in range(N_GENERATIONS):x,y=translateDNA(population)population = np.array( crossover_and_mutaion(population,Crossover_rate=0.8))fitness=get_fitness(population)pop = select(population,fitness)if i == N_GENERATIONS - 1:x1, y1 = translateDNA(pop)sca = ax.scatter(x1, y1, F(x1, y1), c='green', marker='*',linewidths=5)plt.show()plt.pause(0.1)print("迭代:",i)print(population)print_info(population)plt.ioff()plot_3d(ax)【附录】
1.np.argpartition可以取数组中最大的k个值的序号;或者最小k个值的序号
最小3个值的序号: np.argpartition(arr, 3)[:3]
最大3个值的序号: np.argpartition(arr, -3)[-3:]
最小值序号举例如下:
arr = np.array([3, 1, 4, 1, 5, 9, 2, 6]) ---> [3, 1, 4, 1, 5, 9, 2, 6]
indices = np.argpartition(arr, 3) ---> [1 3 6 0 2 4 7 5]
np.argpartition(arr, 3)[:3] -----> array([1, 3, 6])
最大值序号举例如下:
np.argpartition(arr, -3)[-3:] ----> array([4, 7, 5])
2.np.random.choice(a),从数组中选择,可指定概率,可放回/不放回
np.random.choice(a, size=None, replace=True, p=None)
| 参数 | 说明 | 默认值 |
|---|---|---|
a | 输入数组或整数 | 必需 |
size | 输出形状 | None |
replace | 是否允许重复抽样 | True |
p | 每个元素的概率分布 | None |
np.random.choice(data,7,replace=False) --》 array([5, 6, 3, 2, 9, 0, 4])
np.random.choice(data,7,replace=True) -》array([6, 4, 6, 9, 1, 7, 6])
data = np.arange(5) ---》 array([0, 1, 2, 3, 4])
p=[0.1,0.1,0.2,0.3,0.3]
np.random.choice(data, size=8, p=p) ----》 array([3, 1, 4, 4, 2, 1, 4, 3])
