AlphaFold 2 本地部署与安装教程(Linux)
写在前面
蛋白质对生命至关重要,了解其结构有助于理解其功能。通过巨大的实验努力,已测定了约 10万个独特蛋白质的结构,但这仅占已知数十亿种蛋白质序列的一小部分。结构覆盖率的瓶颈在于确定单个蛋白质结构需要数月到数年的艰辛工作。需要精确的计算方法来弥补这一差距,并实现大规模的结构生物信息学。仅根据氨基酸序列预测蛋白质将采取的三维结构——即“蛋白质折叠问题”的结构预测部分——已成为一个重要的开放研究问题超过 50 年。尽管近年来取得了进展,但现有方法在缺乏同源结构时远未达到原子精度。研究人员首次提供了一种能够定期预测蛋白质结构的方法,即使在没有已知类似结构的情况下也能达到原子精度。 验证了基于神经网络模型 AlphaFold 的全新设计版本,在极具挑战性的第 14 届蛋白质结构预测关键评估(CASP14) 15 中,展示了在大多数情况下与实验结构相比具有竞争力的准确性,并大幅超越其他方法。最新版本的 AlphaFold 基于一种新型机器学习方法,该方法将关于蛋白质结构的物理和生物学知识,以及多序列比对,融入深度学习算法的设计中。
本文介绍如何在 Linux 环境下通过 conda + pip 搭建最新版本的 AlphaFold 2.3.2 的运行环境,而不依赖 Docker。虽然AlphaFold 3 (AF3)已经发布,现在可以能通过在线服务使用,但是AF3的开源需要申请比较麻烦,并且单体预测AF2和AF3差异并不大,Alphafold 2依然在被广泛使用。Alphafold 2的使用大部分都是在超算或者集群上。其主要安装方式有两种:docker安装和非docker安装。Deepmind开源代码中提供的是构建docker镜像来运行AF2。由于网络环境差异,在国内完全按照代码中说明一步步来安装是基本不可能的。这里我们使用非docker方式安装和运行AF2,可以顺畅完成整个安装流程,参考alphafold_non_docker。
一、安装教程
本例安装环境如下:
Ubuntu 22.04 LTS # lsb_release -a
CUDA Version: 12.8 # nvidia-smi
NVIDIA L40, 46068 MiB # nvidia-smi --query-gpu=gpu_name,memory.total --format=csv
16T 数据盘1. 下载 AlphaFold 源码
先获取官方源码并进入目录:
git clone https://github.com/deepmind/alphafold.git
cd alphafold
2. 安装 Miniconda(已有忽略)
下载并安装最新版本的 Miniconda(Linux x86_64):
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh bash Miniconda3-latest-Linux-x86_64.sh 安装完成后重新加载环境变量:
source ~/.bashrc3. 创建 conda 环境
创建名为 alphafold 的 conda 环境,并指定 Python 版本为 3.8:
conda create -n af2 python=3.10 -y更新 conda(可选):
conda update -n base conda激活环境:
conda activate af24. 安装依赖包
4.1 安装基础依赖
设置原生依赖(严格用 conda-forge);
conda config --set channel_priority strict
conda config --add channels conda-forge
conda config --add channels bioconda
安装 CUDA Toolkit、cuDNN、OpenMM、HMMER、HHsuite、Kalign、PDBFixer 等:
conda install -y \cudatoolkit=11.8 \"cudnn>=8.6,<9" \openmm=7.7.0 \hmmer=3.3.2 \hhsuite=3.3.0 \kalign2=2.04 \pdbfixer \mock⚠️注意:cudnn=8.6.0 在部分渠道下找不到,推荐用 "cudnn>=8.6,<9"。
4.2 安装 Python 依赖
在项目目录里准备一个 requirements_update.txt(或自己命名),内容如下:
absl-py==1.0.0
biopython==1.79
chex==0.1.6
dm-haiku==0.0.12
dm-tree==0.1.8
docker==5.0.0
immutabledict==2.0.0
ml-collections==0.1.0
pillow==10.1.0# --- 科学栈与 JAX 生态(与 py3.8 兼容)---
numpy==1.24.3
scipy==1.11.1
pandas==1.5.3
ml-dtypes==0.2.0
jax==0.4.25# --- TensorFlow (仅 AF2 的 tf.* 模块用到) ---
tensorflow-gpu==2.11.0
zipp==3.17.0然后安装:
pip install -r requirements_update.txt5. 安装 jaxlib(CUDA 对应版本)
根据 CUDA & cuDNN 选择合适的 wheel(这里是 CUDA 11 + cuDNN 8.6):
pip install --no-cache-dir \"jaxlib==0.4.25+cuda11.cudnn86" \-f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html
6. 下载残基参数文件
AlphaFold 需要下载一个残基参数文件(必要依赖):
#下载残基性质参数到common文件夹
wget -q -P alphafold/common/ https://git.scicore.unibas.ch/schwede/openstructure/-/raw/7102c63615b64735c4941278d92b554ec94415f8/modules/mol/alg/src/stereo_chemical_props.txt7. 应用 OpenMM 补丁(可选)
如果是通过下载压缩文件得到的是老版本的Alphafold v2.3.1(如下),则需要执行这一步骤,最新版是不需要的。
wget https://github.com/deepmind/alphafold/archive/refs/tags/v2.3.1.tar.gz && tar -xzf v2.3.1.tar.gz进入 site-packages 路径,应用 OpenMM 补丁,如果你用的是miniconda,命令如下:
# $alphafold_path 变量设置为 alphafold git repo 目录(绝对路径)
cd ~/miniconda3/envs/af2/lib/python3.8/site-packages/ && patch -p0 < ./docker/openmm.patch如果你用的是 anaconda3 路径,命令改为:
cd ~/anaconda3/envs/af2/lib/python3.8/site-packages/ && patch -p0 < ./docker/openmm.patch8. 环境验证
# 测试 JAX:确认 JAX 能找到 GPU 设备。
python - <<'PY'
import jax
print("JAX:", jax.__version__, "Devices:", jax.devices())
print("Has jax.tree? ->", hasattr(jax, "tree"))
PY
# 输出结果如下:
# JAX: 0.4.25 Devices: [cuda(id=0)]
# Has jax.tree? -> True# 测试 cuDNN:直接尝试加载动态库,确保 CUDA/cuDNN 库能被找到。
python - <<'PY'
import ctypes, os
ctypes.CDLL("libcudnn.so.8")
print("cuDNN OK at:", os.environ.get("CONDA_PREFIX"))
PY
# 输出:cuDNN OK at: /home/username/miniconda3/envs/af2# 测试 OpenMM CUDA:验证 分子动力学 relax 环节依赖的 OpenMM 是否能调用 GPU
python - <<'PY'
import openmm as mm
print("OpenMM platform:", mm.Platform.getPlatformByName('CUDA').getName())
PY
# 输出:OpenMM platform: CUDA# 测试 Haiku Flax 接口:验证 AlphaFold2 模型层用到的 Haiku/Flax 桥接接口是否正常。
python - <<'PY'
from haiku.experimental import flax as hk_flax
print("haiku.experimental.flax OK")
PY
# 输出:haiku.experimental.flax OK最小化 GPU 检测:
python -c "import jax; print(jax.local_devices()[0].platform)" # 应输出 'gpu'
-
gpu→ 说明 JAX 已经正确识别并使用到 NVIDIA GPU。 -
cpu→ 说明 JAX 只找到了 CPU(可能是 CUDA没配好,或者用的是 CPU-only 版本的 JAX)。 -
报错 → 多半是 JAX 版本和 CUDA不匹配。
如果都能输出正确结果 → 环境配置成功!
9. 下载 AlphaFold 所需数据库
AlphaFold 需要多个数据库(UniRef90、MGnify、PDB70、BFD、Uniclust30 等)来完成序列比对和结构预测。下载数据集应该是国内用户安装最大的障碍同时也是最耗时的步骤了,但同时这一步也是最难通过其他办法解决的。整个需要的数据集包括 9 项数据,下载解压后大小~2.7T,可以通过下载的alphafold `scripts`目录下的`download_all_data.sh`进行下载。
注意:保存数据库文件夹需要在alphafold主文件夹以外。alphafold主文件夹可以位于任意位置,保持读写权限即可。
9.1 检查脚本
首先确保 download_db.sh 脚本在 $alphafold_path/scripts/ 目录下,脚本内容如下:
#!/bin/bash
#
# Copyright 2021 DeepMind Technologies Limited
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#
# Downloads and unzips all required data for AlphaFold.
#
# Usage: bash download_all_data.sh /path/to/download/directory
set -eif [[ $# -eq 0 ]]; thenecho "Error: download directory must be provided as an input argument."exit 1
fiif ! command -v aria2c &> /dev/null ; thenecho "Error: aria2c could not be found. Please install aria2c (sudo apt install aria2)."exit 1
fiDOWNLOAD_DIR="$1"
DOWNLOAD_MODE="${2:-full_dbs}" # Default mode to full_dbs.
if [[ "${DOWNLOAD_MODE}" != full_dbs && "${DOWNLOAD_MODE}" != reduced_dbs ]]
thenecho "DOWNLOAD_MODE ${DOWNLOAD_MODE} not recognized."exit 1
fiSCRIPT_DIR="$(dirname "$(realpath "$0")")"echo "Downloading AlphaFold parameters..."
bash "${SCRIPT_DIR}/download_alphafold_params.sh" "${DOWNLOAD_DIR}"if [[ "${DOWNLOAD_MODE}" = reduced_dbs ]] ; thenecho "Downloading Small BFD..."bash "${SCRIPT_DIR}/download_small_bfd.sh" "${DOWNLOAD_DIR}"
elseecho "Downloading BFD..."bash "${SCRIPT_DIR}/download_bfd.sh" "${DOWNLOAD_DIR}"
fiecho "Downloading MGnify..."
bash "${SCRIPT_DIR}/download_mgnify.sh" "${DOWNLOAD_DIR}"echo "Downloading PDB70..."
bash "${SCRIPT_DIR}/download_pdb70.sh" "${DOWNLOAD_DIR}"echo "Downloading PDB mmCIF files..."
bash "${SCRIPT_DIR}/download_pdb_mmcif.sh" "${DOWNLOAD_DIR}"echo "Downloading Uniref30..."
bash "${SCRIPT_DIR}/download_uniref30.sh" "${DOWNLOAD_DIR}"echo "Downloading Uniref90..."
bash "${SCRIPT_DIR}/download_uniref90.sh" "${DOWNLOAD_DIR}"echo "Downloading UniProt..."
bash "${SCRIPT_DIR}/download_uniprot.sh" "${DOWNLOAD_DIR}"echo "Downloading PDB SeqRes..."
bash "${SCRIPT_DIR}/download_pdb_seqres.sh" "${DOWNLOAD_DIR}"echo "All data downloaded."
set -e:任一命令出错就立刻退出(避免“下载一半还继续”的脏状态)。参数校验:必须传入 下载目录(
$1),否则报错退出。依赖检查:必须先安装
aria2c,否则报错退出(这也解释了为啥装了 aria2 下载更快)。
DOWNLOAD_MODE:第二个可选参数,默认为full_dbs;也可传reduced_dbs(精简库)。
SCRIPT_DIR:脚本自身目录的绝对路径,后面会用它去调用同目录下的其他分脚本(如download_bfd.sh等)。
9.2 安装 aria2
安装 aria2 能显著提升下载效率,尤其是像 BFD (~1.7TB) 这种超大数据库。必须先安装 aria2c,否则脚本就会报错退出。
sudo apt install aria2# 安装成功后可以查看版本信息
aria2c -v9.3 加速下载配置(可选,但推荐)
给 aria2c 套一层“加速 wrapper”,让所有脚本里调用到的 aria2c 自动带上多线程/断点续传/预分配等参数。
1) 在你的家目录下创建 ~/bin,作为个人可执行文件夹;
mkdir -p ~/bin2)在 ~/bin 里新建一个名为 aria2c 的可执行脚本(和系统的同名),并写入以下内容
cat > ~/bin/aria2c <<'EOF'
#!/usr/bin/env bashexec /usr/bin/aria2c \--max-connection-per-server=16 \--split=16 \--min-split-size=1M \--continue=true \--retry-wait=5 \--timeout=60 \--file-allocation=falloc \--disk-cache=64M \--summary-interval=60 \"$@"
EOF这表示:凡是调用 aria2c,实际都会转发到系统的 /usr/bin/aria2c,但自动带上一组加速/稳定参数,最后把原本命令行的其他参数 "$@" 原样拼上。
参数含义:
-
--max-connection-per-server=16:同一服务器最多 16 个连接(CN 上限)。 -
--split=16:单个文件最多拆成 16 段并发下载。 -
--min-split-size=1M:每段至少 1MB,避免过度分片。 -
--continue=true:断点续传。 -
--retry-wait=5/--timeout=60:失败重试等待与超时设置。 -
--file-allocation=falloc:快速预分配文件空间(ext4/xfs 等很好用)。 -
--disk-cache=64M:磁盘缓存 64MB,提高落盘效率。 -
--summary-interval=60:每 60 秒汇总一次速度摘要。
3)赋予文件可执行权限
chmod +x ~/bin/aria2c4)把 ~/bin 放到 PATH 前面,这样当脚本调用 aria2c 时,会先命中个人目录下的这个“同名 wrapper”
export PATH="$HOME/bin:$PATH"# 小提示:想永久生效,可把上面这行写进 ~/.bashrc或者 ~/.zshrc,然后 source ~/.bashrc 或者 source ~/.zshrc。5)验证是否生效
type -a aria2c # 应显示先命中 ~/bin/aria2c
"""
原本应该显示以下内容:aria2c is /usr/bin/aria2caria2c is /bin/aria2c现在显示多了个人用户下的aria2caria2c is /home/username/bin/aria2caria2c is /usr/bin/aria2caria2c is /bin/aria2c
"""which aria2c
"""
原本显示:/usr/bin/aria2c
设置之后显示:/home/username/bin/aria2c
"""aria2c -v # 能正常显示版本"""
aria2 version 1.36.0
Copyright (C) 2006, 2019 Tatsuhiro TsujikawaThis program is free software; you can redistribute it and/or modify
it under the terms of the GNU General Public License as published by
the Free Software Foundation; either version 2 of the License, or
(at your option) any later version.This program is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
GNU General Public License for more details.** Configuration **
Enabled Features: Async DNS, BitTorrent, Firefox3 Cookie, GZip, HTTPS, Message Digest, Metalink, XML-RPC, SFTP
Hash Algorithms: sha-1, sha-224, sha-256, sha-384, sha-512, md5, adler32
Libraries: zlib/1.2.11 libxml2/2.9.12 sqlite3/3.36.0 GnuTLS/3.7.1 nettle GMP/6.2.1 c-ares/1.17.1 libssh2/1.10.0
Compiler: gcc 11.2.0built by x86_64-pc-linux-gnuon Aug 21 2021 15:26:28
System: Linux 5.15.0-25-generic #25-Ubuntu SMP Wed Mar 30 15:54:22 UTC 2022 x86_64Report bugs to https://github.com/aria2/aria2/issues
Visit https://aria2.github.io/
"""6)如果下载完之后不需要的话,可以选择彻底删除该文件配置
rm -f ~/bin/aria2c
# 或者把 PATH 恢复(重新登录 shell 或 source ~/.bashrc)
9.4 运行下载脚本
1)下载数据库示例
# 1) 下载“全量数据库”(科研推荐,~2TB+ 磁盘)
bash scripts/download_all_data.sh /home/you/af2_data# 2) 下载“精简数据库”(测试/空间紧张,~400–600GB)
bash scripts/download_all_data.sh /home/you/af2_data reduced_dbs2)下载全部的数据库推荐以下运行方式(带日志、后台跑),这里注意自己服务器文件目录的修改,如果空间不够需要下载精简数据库的话,可以指令后面添加关键词 reduced_dbs 即可,注意这里路径修改为自己服务器的路径
mkdir -p /data1/database/af2_database # 根据自己服务器的情况再大磁盘创建目录,数据库安装路径可以是任意位置
cd /data1/database/af2_database
nohup bash /data1/username/alphafold-2.3.2/scripts/download_all_data.sh /data1/database/af2_database > download.log 2> download_error.log &
tail -f download.log # 实时看进度# 注意:如果关闭终之后下载任务被杀后台的话,可以执行以下指令
nohup setsid bash /data1/username/alphafold-2.3.2/scripts/download_all_data.sh /data1/database/af2_database > /data1/database/af2_database/download.log 2>&1 < /dev/null &# 或者使用tmux创建一个“虚拟终端”,即使你关掉 SSH/终端,里面的任务还在运行
tmux new -s af2 -d 'bash /data1/username/alphafold-2.3.2/scripts/download_all_data.sh /data1/database/af2_database > download.log 2> download_error.log'
tmux ls # 查看是否在跑,会显示af2会话
tmux attach -t af2 # 随时再连上看进度;Ctrl+b 然后 d 脱离使用nohup在后台运行 AlphaFold 数据库下载脚本,把标准输出写入 download.log,错误信息写入 download_error.log,即使退出终端也不会中断下载。
3)如果觉得下载比较慢可以返回 9.3 配置加速下载,配置好之后需要先停掉当前的 aria2c,然后在执行第 2)步下载
pkill -f aria2c
# or
kill -9 <PID># 确认没有残留
ps -ef | grep aria2c4)下载速度比较
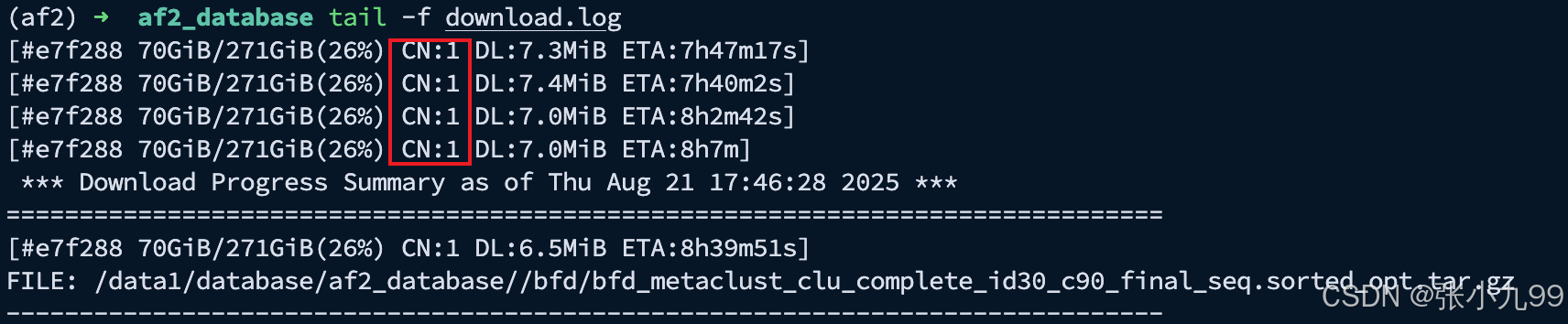
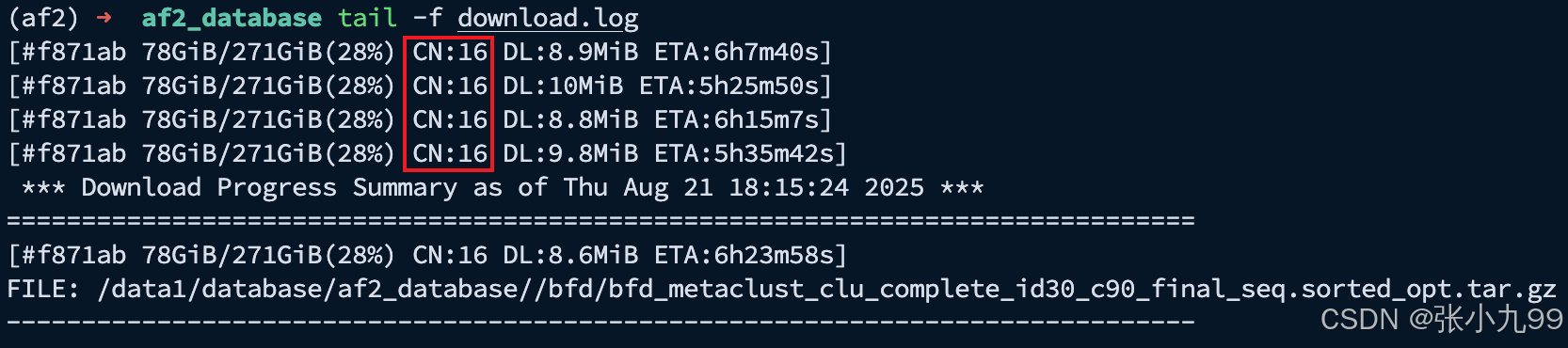
可以发现修改的配置生效了CN:16从由 1 -> 16,CN = Connection Number(当前并发连接数),表示 aria2c 这个下载任务同时开了多少条 TCP 连接 去同一个服务器请求不同的数据分片。同时可以发现预计剩余时间ETA也会缩短,并且下载速度也加快了。
9.5 下载可能遇到的问题
问题一:下载pdb_mmcif数据库时报错
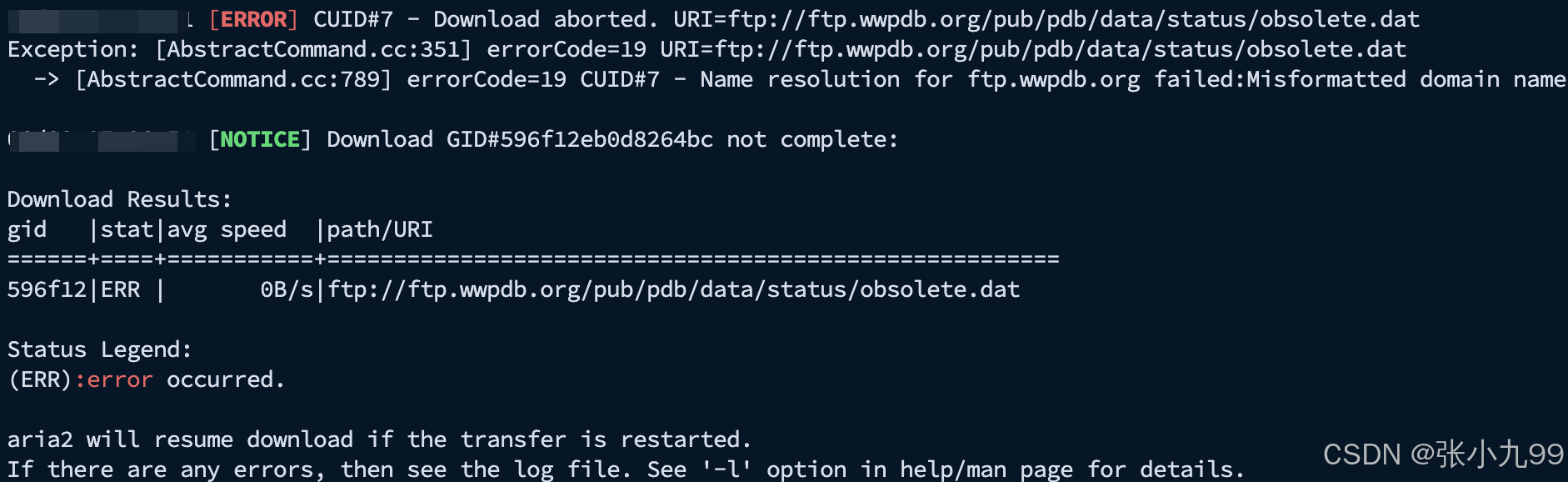
原因:
看日志是 aria2c 在拉取 ftp://ftp.wwpdb.org/pub/pdb/data/status/obsolete.dat 时报错(ERR)。核心原因很可能是:wwPDB 已经弃用 FTP 下载,要求改用 HTTPS(files.wwpdb.org)或 RSYNC,所以老脚本里指向 ftp.wwpdb.org 的链接会失败。
解决方案:
1)打开你用来下 AF2 数据库的脚本 alphafold-2.3.2/scripts/download_pdb_mmcif.sh;
2)修改脚本中最后一行;
# 原始内容
aria2c "ftp://ftp.wwpdb.org/pub/pdb/data/status/obsolete.dat" --dir="${ROOT_DIR}"# 修改为一下内容
aria2c "https://files.wwpdb.org/pub/pdb/data/status/obsolete.dat" --dir="${ROOT_DIR}"3)重新运行下载(aria2c 会自动断点续传);
问题二:下载中断后重新运行脚本时,params文件会重新下载
原因:
download_all_data.sh 在下载 AlphaFold 参数文件 (alphafold_params_2022-12-06.tar) 之后,脚本会 自动解压 这个 .tar 包。解压后,里面的内容就是模型参数文件params_model_x.npz 系列文件(包括 multimer、ptm 等)。为了节省磁盘空间,脚本默认会删除原始 .tar 压缩包,只保留解压后的 .npz 文件。aria2c 本身是支持断点续传的,机制是脚本在调用 aria2c 下载参数文件时,默认会检查目标路径是否已有完整文件。如果之前下载 未完成(比如 .tar.aria2 还在),aria2c 会自动续传。虽然之前已经下载完成,但解压后 .tar 被删除,下次再运行时,它就认为参数文件“不存在”,于是又重新下载并解压。
解决方案一:
1)打开你用来下 AF2 数据库的脚本 alphafold-2.3.2/scripts/download_alphafold_params.sh;
2)将最后一行删除参数文件压缩包.tar的命令注释掉即可
# rm "${ROOT_DIR}/${BASENAME}"3)重新运行下载命令后开始下载后,以后再遇到中断重新下载就不会再重复下载参数文件了,节省下载了时间;
类似的,后续下载其他数据库也会遇到这个问题,比如bfd
解决方案二:
修改数据库下载脚本download_bfd.sh,在里面新增文件存在性判断(其他数据库也可以用类似的解决方案),如下所示:
#!/bin/bash
#
# Copyright 2021 DeepMind Technologies Limited
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#
# Downloads and unzips the BFD database for AlphaFold.
#
# Usage: bash download_bfd.sh /path/to/download/directory
set -eif [[ $# -eq 0 ]]; thenecho "Error: download directory must be provided as an input argument."exit 1
fiif ! command -v aria2c &> /dev/null ; thenecho "Error: aria2c could not be found. Please install aria2c (sudo apt install aria2)."exit 1
fiDOWNLOAD_DIR="$1"
ROOT_DIR="${DOWNLOAD_DIR}/bfd"
# Mirror of:
# https://bfd.mmseqs.com/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt.tar.gz.
SOURCE_URL="https://storage.googleapis.com/alphafold-databases/casp14_versions/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt.tar.gz"
BASENAME=$(basename "${SOURCE_URL}")# 已解压后的关键文件前缀
BFD_PREFIX="bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt"# 如果 6 个核心文件都存在,就跳过下载
if [ -s "${ROOT_DIR}/${BFD_PREFIX}_a3m.ffdata" ] && [ -s "${ROOT_DIR}/${BFD_PREFIX}_a3m.ffindex" ] \&& [ -s "${ROOT_DIR}/${BFD_PREFIX}_hhm.ffdata" ] && [ -s "${ROOT_DIR}/${BFD_PREFIX}_hhm.ffindex" ] \&& [ -s "${ROOT_DIR}/${BFD_PREFIX}_cs219.ffdata" ] && [ -s "${ROOT_DIR}/${BFD_PREFIX}_cs219.ffindex" ]; thenecho "✅ BFD already present in ${ROOT_DIR}, skipping download."exit 0
fimkdir --parents "${ROOT_DIR}"
aria2c "${SOURCE_URL}" --dir="${ROOT_DIR}"
tar --extract --verbose --file="${ROOT_DIR}/${BASENAME}" \--directory="${ROOT_DIR}"
rm "${ROOT_DIR}/${BASENAME}"54,1 Bot9.6 下载完成目录结构
tail -f download.log上述指令查看下载日志文件,如果显示以下信息,说明最后一个文件 pdb_seqres 也下载完成,所有的数据库都完成下载;
08/25 04:28:09 [NOTICE] Download complete: /data1/database/af2_database/pdb_seqres/pdb_seqres.txtDownload Results:
gid |stat|avg speed |path/URI
======+====+===========+=======================================================
d9d850|OK | 8.2MiB/s|/data1/database/af2_database/pdb_seqres/pdb_seqres.txtStatus Legend:
(OK):download completed.
All data downloaded.tree -L 2 /data1/database/af2_database可以用以上指令查看下载的目录,下载完成后,你的目录结构类似如下,但是不同时间下载可能数据库会有更新,大小会有些许差异,以下的下载日期为August 2025,可做参考;
/data1/database/af2_database
├── bfd # 6 files ~ 1.8 TB (download: 271.6 GB)
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_a3m.ffdata
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_a3m.ffindex
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_cs219.ffdata
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_cs219.ffindex
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_hhm.ffdata
│ └── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_hhm.ffindex
├── mgnify # 1 file ~ 120 GB (download: 67 GB)
│ └── mgy_clusters_2022_05.fa
├── params # 16 files 5 ~ 5.3 GB (download: 5.3 GB): CASP14 models,5 pTM models,5 AlphaFold-Multimer models,LICENSE
│ ├── LICENSE
│ ├── params_model_1_multimer_v3.npz
│ ├── params_model_1.npz
│ ├── params_model_1_ptm.npz
│ ├── params_model_2_multimer_v3.npz
│ ├── params_model_2.npz
│ ├── params_model_2_ptm.npz
│ ├── params_model_3_multimer_v3.npz
│ ├── params_model_3.npz
│ ├── params_model_3_ptm.npz
│ ├── params_model_4_multimer_v3.npz
│ ├── params_model_4.npz
│ ├── params_model_4_ptm.npz
│ ├── params_model_5_multimer_v3.npz
│ ├── params_model_5.npz
│ └── params_model_5_ptm.npz
├── pdb70 # 9 files ~ 56 GB (download: 19.5 GB)
│ ├── md5sum
│ ├── pdb70_a3m.ffdata
│ ├── pdb70_a3m.ffindex
│ ├── pdb70_clu.tsv
│ ├── pdb70_cs219.ffdata
│ ├── pdb70_cs219.ffindex
│ ├── pdb70_hhm.ffdata
│ ├── pdb70_hhm.ffindex
│ └── pdb_filter.dat
├── pdb_mmcif # About 240,682 .cif files ~ 324 GB
│ ├── mmcif_files
│ └── obsolete.dat
├── pdb_seqres # 1 file ~ 0.3 GB (download: 0.3 GB)
│ └── pdb_seqres.txt
├── uniprot # 1 file ~ 116 GB
│ └── uniprot.fasta
├── uniref30 # 1 file ~ 206 GB (download: 52.5 GB)
│ ├── UniRef30_2021_03_a3m.ffdata
│ ├── UniRef30_2021_03_a3m.ffindex
│ ├── UniRef30_2021_03_cs219.ffdata
│ ├── UniRef30_2021_03_cs219.ffindex
│ ├── UniRef30_2021_03_hhm.ffdata
│ ├── UniRef30_2021_03_hhm.ffindex
│ └── UniRef30_2021_03.md5sums
└── uniref90 # 1 file ~ 92 GB└── uniref90.fasta 另外,如果网络受限,或需要定制,可以参考官方链接手动下载:
👉 AlphaFold 数据库下载说明
9.7 修改数据库权限
建议只读(防止误删/误改),但 owner 要能写:
# 所有子目录与文件设置为 755(目录可进入,文件可读)
sudo chmod -R 755 /data1/database/af2_database
这样:
-
你(owner)有读写权限
-
组用户、其他用户只有读和进入权限
-
MSA 工具能正常读取
到目前未知,Alphafold 2 的安装部分就结束了,可以执行下面的脚本进行测试以及结构预测。
二、使用教程
1. 运行测试脚本
运行以下测试脚本,
python run_alphafold_test.py 显示以下信息,表明 AlphaFold2 的环境安装和运行测试完全成功 ✅
Running tests under Python 3.10.18: /home/username/miniconda3/envs/af2/bin/python
[ RUN ] RunAlphafoldTest.test_end_to_end_no_relax
I0826 17:09:17.350987 140467440940864 run_alphafold.py:246] Predicting test
I0826 17:09:17.351292 140467440940864 run_alphafold.py:277] Running model model1 on test
I0826 17:09:17.351357 140467440940864 run_alphafold.py:289] Total JAX model model1 on test predict time (includes compilation time, see --benchmark): 0.0s
I0826 17:09:17.376969 140467440940864 run_alphafold.py:415] Final timings for test: {'features': 2.9802322387695312e-05, 'process_features_model1': 1.52587890625e-05, 'predict_and_compile_model1': 1.2874603271484375e-05}
[ OK ] RunAlphafoldTest.test_end_to_end_no_relax
[ RUN ] RunAlphafoldTest.test_end_to_end_relax
I0826 17:09:17.378478 140467440940864 run_alphafold.py:246] Predicting test
I0826 17:09:17.378607 140467440940864 run_alphafold.py:277] Running model model1 on test
I0826 17:09:17.378669 140467440940864 run_alphafold.py:289] Total JAX model model1 on test predict time (includes compilation time, see --benchmark): 0.0s
I0826 17:09:17.414821 140467440940864 run_alphafold.py:415] Final timings for test: {'features': 2.193450927734375e-05, 'process_features_model1': 1.6450881958007812e-05, 'predict_and_compile_model1': 1.0967254638671875e-05, 'relax_model1': 2.765655517578125e-05}
[ OK ] RunAlphafoldTest.test_end_to_end_relax
----------------------------------------------------------------------
Ran 2 tests in 0.066sOK说明:
核心依赖 (JAX, Haiku, OpenMM, TensorFlow, CUDA/cuDNN) 已经全部正常工作;
Relaxation 步骤(OpenMM AMBER 最小化)也跑通了;
可能会看到的
libnvinfer.so.7/TensorRT警告,可以无视(除非你想用 TensorRT 加速推理,但 AF2 默认不用)。
2. 运行测试预测
(1)示例命令(monomer 模式,使用全数据库):
python run_alphafold.py \--fasta_paths=/path/to/query.fasta \--output_dir=/path/to/output \--data_dir=/data1/database/af2_database \--uniref90_database_path=/data1/database/af2_database/uniref90/uniref90.fasta \--mgnify_database_path=/data1/database/af2_database/mgnify/mgy_clusters_2022_05.fa \--template_mmcif_dir=/data1/database/af2_database/pdb_mmcif/mmcif_files \--obsolete_pdbs_path=/data1/database/af2_database/pdb_mmcif/obsolete.dat \--pdb70_database_path=/data1/database/af2_database/pdb70/pdb70 \--uniref30_database_path=/data1/database/af2_database/uniref30/UniRef30_2021_03 \--bfd_database_path=/data1/database/af2_database/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \--max_template_date=2025-07-31 \--models_to_relax=best \--use_gpu_relax=true \--db_preset=full_dbs \--model_preset=monomer
--fasta_paths=/path/to/query.fasta
要预测的序列文件。支持单条或多条序列(多条时会按 multimer 处理,但你这里搭配的是 monomer 预设)。多个 FASTA 也可以用逗号分隔传入。
--output_dir=/path/to/output
结果输出根目录。AlphaFold 会在这里新建一个以 FASTA 基名命名的子目录,保存特征、模型输出、排名、PDB/CIF 和日志等。
--data_dir=/data1/database/af2_database
AlphaFold 参数与若干索引文件的根目录(不是所有数据库都从这里自动推断,所以下面仍需逐一指定路径)。
--uniref90_database_path
给 JackHMMER 用的 UniRef90 蛋白库,做深度同源检索(MSA)。
--mgnify_database_path
MGnify 宏基因组序列聚类库,弥补常规数据库覆盖不足。
--uniref30_database_path
HHblits 用的 UniRef30(foldseek/hhblits 的 cs219/hhm/a3m 索引都在这个前缀下)。
--bfd_database_path
BFD 超大序列数据库(或其 metaclust 版本),配合 HHblits 进一步扩展 MSA。
--template_mmcif_dir
模板结构库(PDB 的 mmCIF 扁平化目录,每个条目形如7abc.cif)。
模板命中由 HHsearch(或 multimer 时的 hmmsearch)在这里取坐标。
--obsolete_pdbs_path=
PDB 过时条目映射表。用于把已废弃的 PDB ID 映射到替代条目,避免用到过时结构。
--pdb70_database_path=
PDB70(序列聚类版 PDB)给 HHsearch 做模板搜索时使用(仅 monomer 模型需要)。
--max_template_date=2025-07-31
模板的最晚发布日期(知识截止)。设置它可以避免“信息泄露”(例如做历史集评测时不允许使用未来的模板)。
实际生产中若只追求最优结构,可以设为极大值(如9999-12-31)以启用所有模板。也可以选一个 你数据库下载日期之前 的 cutoff,比如如果数据库是2025-08下的,就用--max_template_date=2025-07-31
--models_to_relax=best
AMBER Relax(几何优化)的范围:
none:不做 relax(最快,但可能有局部几何问题)
best:只对排名第 1 的结果 relax(折中,常用设置)
all:对所有模型/采样结果都 relax(最稳,但最慢)
--use_gpu_relax=true
若 GPU 上装有支持的 OpenMM/CUDA,Relax 用 GPU 跑会快很多。无法用 GPU 时设false。
--db_preset=full_dbs
MSA 数据库预设:
full_dbs:用 UniRef90 + MGnify + BFD + UniRef30(准确度高,耗时长)
reduced_dbs:用 Small BFD 替代 BFD(快,但 MSA 变浅,精度略降)
--model_preset=monomer
模型预设:
monomer:单体模型(默认)
monomer_ptm:使用 pTM 头进行微调的原始 CASP14 模型,提供了成对置信度测量。其准确度略低于普通单体模型。
monomer_casp14:更强的集成/测试配置(更慢)
multimer:复合体模型(需要不同的数据库组合与模板搜索器)
(2)运行后,输出目录中会有:
-
ranking_debug.json:5 个模型得分排名
{"plddts": {"model_1_pred_0": 95.41898139620274,"model_2_pred_0": 95.57812625885195,"model_3_pred_0": 95.47367863427417,"model_4_pred_0": 95.19971885973155,"model_5_pred_0": 94.62560287140026},"order": ["model_2_pred_0","model_3_pred_0","model_1_pred_0","model_4_pred_0","model_5_pred_0"]
}-
confidence_model_X_pred_0.json(逐残基的 pLDDT)
{"residueNumber":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324,325,326,327,328,329,330,331,332,333,334,335,336],"confidenceScore":[82.73,87.38,86.17,91.02,92.98,93.03,92.78,93.8,93.58,92.29,91.7,92.32,95.55,95.57,94.29,96.53,96.83,98.07,98.24,98.3,98.42,98.3,98.34,98.18,97.88,95.7,95.35,96.77,97.7,97.74,98.2,98.45,98.71,98.61,98.36,97.86,98.12,96.83,95.44,96.75,95.6,96.74,97.02,97.75,97.91,97.55,98.05,97.76,96.94,97.84,97.75,97.43,97.24,96.9,96.33,97.04,96.59,94.89,94.64,95.72,94.13,94.66,94.93,92.5,92.47,95.01,96.8,98.12,98.46,98.36,98.13,95.64,93.65,80.04,85.79,95.82,96.92,97.43,97.83,98.25,98.42,98.38,97.89,97.54,97.5,96.97,96.18,96.2,95.93,97.37,98.01,98.6,98.42,98.36,97.54,94.28,91.14,90.29,84.94,84.66,86.31,90.0,94.06,92.9,94.03,95.64,96.43,97.51,97.3,97.24,97.82,97.69,98.32,98.12,97.64,97.32,96.5,95.89,94.23,90.34,94.05,94.27,91.5,88.36,89.09,89.91,94.22,95.1,93.84,95.32,97.1,97.44,96.03,97.18,97.64,95.82,96.7,97.46,96.86,91.72,92.74,96.73,97.49,97.48,96.97,95.83,95.41,93.43,94.48,96.64,97.19,98.01,97.59,97.88,98.22,98.08,98.37,98.45,97.14,96.95,97.74,98.2,97.47,97.61,98.1,98.04,97.61,97.58,97.77,97.15,96.89,96.27,97.01,97.33,97.67,98.69,98.71,98.7,98.83,98.77,98.41,97.46,96.43,93.59,94.08,94.93,97.26,97.88,97.66,97.49,98.17,98.53,98.3,97.98,98.46,98.58,98.23,98.14,98.25,98.34,97.94,97.55,98.55,98.6,98.56,98.35,97.97,98.04,96.71,96.51,95.08,91.83,89.02,89.93,91.42,91.75,91.17,89.64,86.79,83.7,83.32,80.88,84.44,85.53,90.49,91.94,93.07,89.37,86.72,91.75,86.65,83.71,83.74,87.18,90.86,90.85,92.48,92.56,92.75,93.86,92.39,94.03,94.12,96.4,97.64,95.51,97.39,97.95,97.45,97.92,97.31,96.91,97.42,98.37,98.36,98.03,98.18,98.61,98.36,97.81,97.53,98.04,96.0,95.37,96.06,93.91,95.85,96.6,97.75,96.96,96.96,97.34,98.41,98.48,98.56,98.74,98.79,98.74,98.68,98.78,98.52,98.48,98.18,97.79,97.42,97.2,97.66,97.13,94.96,95.88,98.24,98.56,98.79,98.83,98.84,98.63,98.4,98.09,97.63,97.24,96.43,94.81,92.87,93.94,94.35,92.26,92.42,93.51,95.38,95.46,95.15,94.44,95.08,94.62,95.99,96.14,94.77,94.99,97.79,97.8,97.7,96.5,97.1,98.25,97.77,98.15,97.61,97.3,97.68,97.06,96.8,96.28,88.46,81.22,78.23,60.58],"confidenceCategory":["M","M","M","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","M","M","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","M","M","M","M","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","M","M","M","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","M","M","H","H","H","M","M","M","M","M","M","M","H","H","H","M","M","H","M","M","M","M","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","H","M","M","M","L"]}-
timings.json(各阶段耗时统计)
{"features": 1411.5304100513458,"process_features_model_1_pred_0": 2.524388551712036,"predict_and_compile_model_1_pred_0": 70.85654354095459,"process_features_model_2_pred_0": 1.9797868728637695,"predict_and_compile_model_2_pred_0": 64.33179903030396,"process_features_model_3_pred_0": 1.7526803016662598,"predict_and_compile_model_3_pred_0": 49.07119393348694,"process_features_model_4_pred_0": 1.7196385860443115,"predict_and_compile_model_4_pred_0": 47.63682198524475,"process_features_model_5_pred_0": 1.5123274326324463,"predict_and_compile_model_5_pred_0": 46.46093392372131,"relax_model_2_pred_0": 25.11619257926941
}-
ranked_0.pdb~ranked_4.pdb(按预测质量排序的结构) -
relaxed_model_X.pdb(AMBER 最小化后的结构)
(3)relaxed_model_1_pred_0.pdb 为经 OpenMM Amber relaxation 后的结构,Relaxation 主要用于消除不合理的原子冲突和几何约束。文件中第 11 列(B-factor 列)为 pLDDT 分数。Relaxation 使用 OpenMM 调用 AMBER ff14SB 力场,可在 GPU 上加速运行。
(4)输出目录结构如下:
├── confidence_model_1_pred_0.json
├── confidence_model_2_pred_0.json
├── confidence_model_3_pred_0.json
├── confidence_model_4_pred_0.json
├── confidence_model_5_pred_0.json
├── features.pkl
├── msas
├── ranked_0.cif
├── ranked_0.pdb
├── ranked_1.cif
├── ranked_1.pdb
├── ranked_2.cif
├── ranked_2.pdb
├── ranked_3.cif
├── ranked_3.pdb
├── ranked_4.cif
├── ranked_4.pdb
├── ranking_debug.json
├── relaxed_model_2_pred_0.cif
├── relaxed_model_2_pred_0.pdb
├── relax_metrics.json
├── result_model_1_pred_0.pkl
├── result_model_2_pred_0.pkl
├── result_model_3_pred_0.pkl
├── result_model_4_pred_0.pkl
├── result_model_5_pred_0.pkl
├── timings.json
├── unrelaxed_model_1_pred_0.cif
├── unrelaxed_model_1_pred_0.pdb
├── unrelaxed_model_2_pred_0.cif
├── unrelaxed_model_2_pred_0.pdb
├── unrelaxed_model_3_pred_0.cif
├── unrelaxed_model_3_pred_0.pdb
├── unrelaxed_model_4_pred_0.cif
├── unrelaxed_model_4_pred_0.pdb
├── unrelaxed_model_5_pred_0.cif
└── unrelaxed_model_5_pred_0.pdb3. Shell 脚本封装
新建一个run_af2.sh的文件,将下面代码本复制进去,修改路径配置,随后就可以很方便的执行该命令进行单个结构预测了:
# 运行run_af2.sh进行单条序列的结构预测
bash run_af2.sh test.fasta ./test 0
# nohup后台稳定运行,即使退出终端也能继续跑
nohup bash run_af2.sh test.fasta ./test 0 > run.log 2>&1 &这里一共有三个参数,分别为输入fasta文件的路径、输出文件路径、显卡ID;
#!/bin/bash
set -euo pipefail # bash 脚本的安全模式
# 用法: bash run_one_af2.sh input.fasta output_dir gpu_idif [[ $# -ne 3 ]]; then # 如果参数不是 3 个,就提示用法并退出,避免少传或多传echo "Usage: $0 <input.fasta> <output_dir> <gpu_id>"exit 1
fi# 输入参数
FASTA=$1
OUTDIR=$2
GPU_ID=$3if [[ ! -f "$FASTA" ]]; then # 这里检查输入的 fasta 文件路径是否有效,如果没找到,就报错并退出echo "ERROR: FASTA not found: $FASTA"exit 2
fi# 初始化 conda
source ~/miniconda3/etc/profile.d/conda.sh
conda activate af2# GPU 控制
export CUDA_VISIBLE_DEVICES=$GPU_ID# 显存策略(可选)
export XLA_PYTHON_CLIENT_PREALLOCATE=true
export XLA_PYTHON_CLIENT_MEM_FRACTION=0.95
export XLA_PYTHON_CLIENT_ALLOCATOR=platform# 限制CPU线程(可选,推荐)
export OMP_NUM_THREADS=1
export OPENBLAS_NUM_THREADS=1
export MKL_NUM_THREADS=1
export NUMEXPR_NUM_THREADS=1# 路径配置
RUN_AF="/data1/username/alphafold/run_alphafold.py"
DATA_DIR="/data1/database/af2_database"
UNIREF90="${DATA_DIR}/uniref90/uniref90.fasta"
MGNIFY="${DATA_DIR}/mgnify/mgy_clusters_2022_05.fa"
PDB_MMCIF_DIR="${DATA_DIR}/pdb_mmcif/mmcif_files"
PDB_OBSOLETE="${DATA_DIR}/pdb_mmcif/obsolete.dat"
PDB70="${DATA_DIR}/pdb70/pdb70"
UNIREF30="${DATA_DIR}/uniref30/UniRef30_2021_03"
BFD="${DATA_DIR}/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt"# 关键路径检查
[[ -f "$RUN_AF" ]] || { echo "ERROR: run_alphafold.py not found: $RUN_AF"; exit 3; }
[[ -d "$DATA_DIR" ]] || { echo "ERROR: DATA_DIR not found: $DATA_DIR"; exit 4; }# AF2 参数
MAX_TEMPLATE_DATE="2025-07-31"
DB_PRESET="full_dbs"
MODEL_PRESET="monomer"
MODELS_TO_RELAX="best"
USE_GPU_RELAX="true"# 日志文件
LOGFILE="${OUTDIR}/af2.log"
mkdir -p "${OUTDIR}"
echo "[$(date)] Run AF2 on GPU ${GPU_ID}" | tee -a "${LOGFILE}"# run_alphafold
python "${RUN_AF}" \--fasta_paths="${FASTA}" \--output_dir="${OUTDIR}" \--data_dir="${DATA_DIR}" \--uniref90_database_path="${UNIREF90}" \--mgnify_database_path="${MGNIFY}" \--template_mmcif_dir="${PDB_MMCIF_DIR}" \--obsolete_pdbs_path="${PDB_OBSOLETE}" \--pdb70_database_path="${PDB70}" \--uniref30_database_path="${UNIREF30}" \--bfd_database_path="${BFD}" \--max_template_date="${MAX_TEMPLATE_DATE}" \--models_to_relax="${MODELS_TO_RELAX}" \--use_gpu_relax="${USE_GPU_RELAX}" \--db_preset="${DB_PRESET}" \--model_preset="${MODEL_PRESET}" \>> "${LOGFILE}" 2>&1echo "[$(date)] Finished ${FASTA}" | tee -a "${LOGFILE}"三、可视化展示
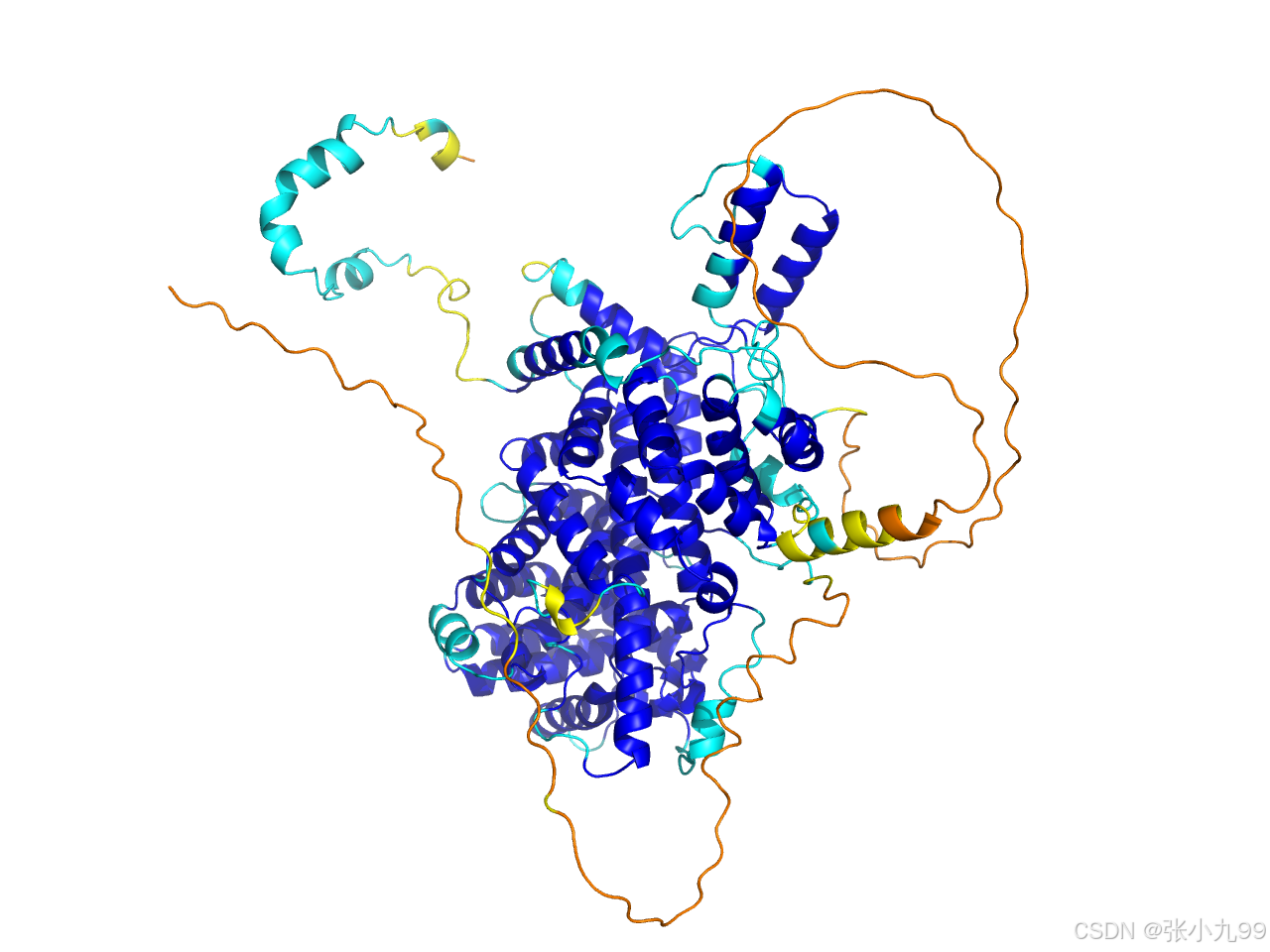
在使用 AlphaFold2 预测得到蛋白质结构之后,我们通常希望快速评估预测的可信度。AlphaFold 在结构文件中已经内置了 pLDDT 分数(每个残基的预测置信度),而利用 PyMOL 插件就可以直观地根据 pLDDT 给结构上色,高置信度和低置信度区域一目了然。
1. 准备pdb结构
将 AlphaFold2 输出的结构文件(如 ranked_0.pdb);
2. 加载扩展插件
在 PyMOL 的命令行窗口中运行以下命令,这一步会从 GitHub 下载并加载 coloraf.py 脚本。
run https://raw.githubusercontent.com/cbalbin-bio/pymol-color-alphafold/master/coloraf.py
3. 载入预测结构
在 PyMOL 中加载你的 AlphaFold2 预测结果,例如:
load ranked_0.pdb, my_model
其中:
-
ranked_0.pdb是 AlphaFold 输出的预测结构文件 -
my_model是在 PyMOL 中的对象名称,你可以根据需要修改
4. 根据 pLDDT 给结构上色
在 PyMOL 命令行中调用扩展:
coloraf my_model插件会自动识别结构文件中的 pLDDT 分数(存储在 B-factor 列),并使用以下默认配色方案上色:
-
蓝色 (高 pLDDT ≥ 90):预测可信度很高
-
青色 (70–90):预测可信度良好
-
黄色 (50–70):预测可信度偏低
-
橙色 (≤ 50):预测可信度很差
这样即可在 PyMOL 中直观查看结构的可信度分布。
总结
-
Python 建议用 3.8–3.10,不要用 3.11+。
-
JAX/JAXLIB 与 CUDA/cuDNN 的对应关系非常重要,必须安装匹配版本。
-
Monomer 模式输出 pLDDT。
-
ranked_0.pdb就是排序最佳的最终模型。
参考文档:
https://github.com/google-deepmind/alphafold/
https://github.com/kalininalab/alphafold_non_docker