MUVERA:让RAG系统中的多向量检索像单向量一样高效
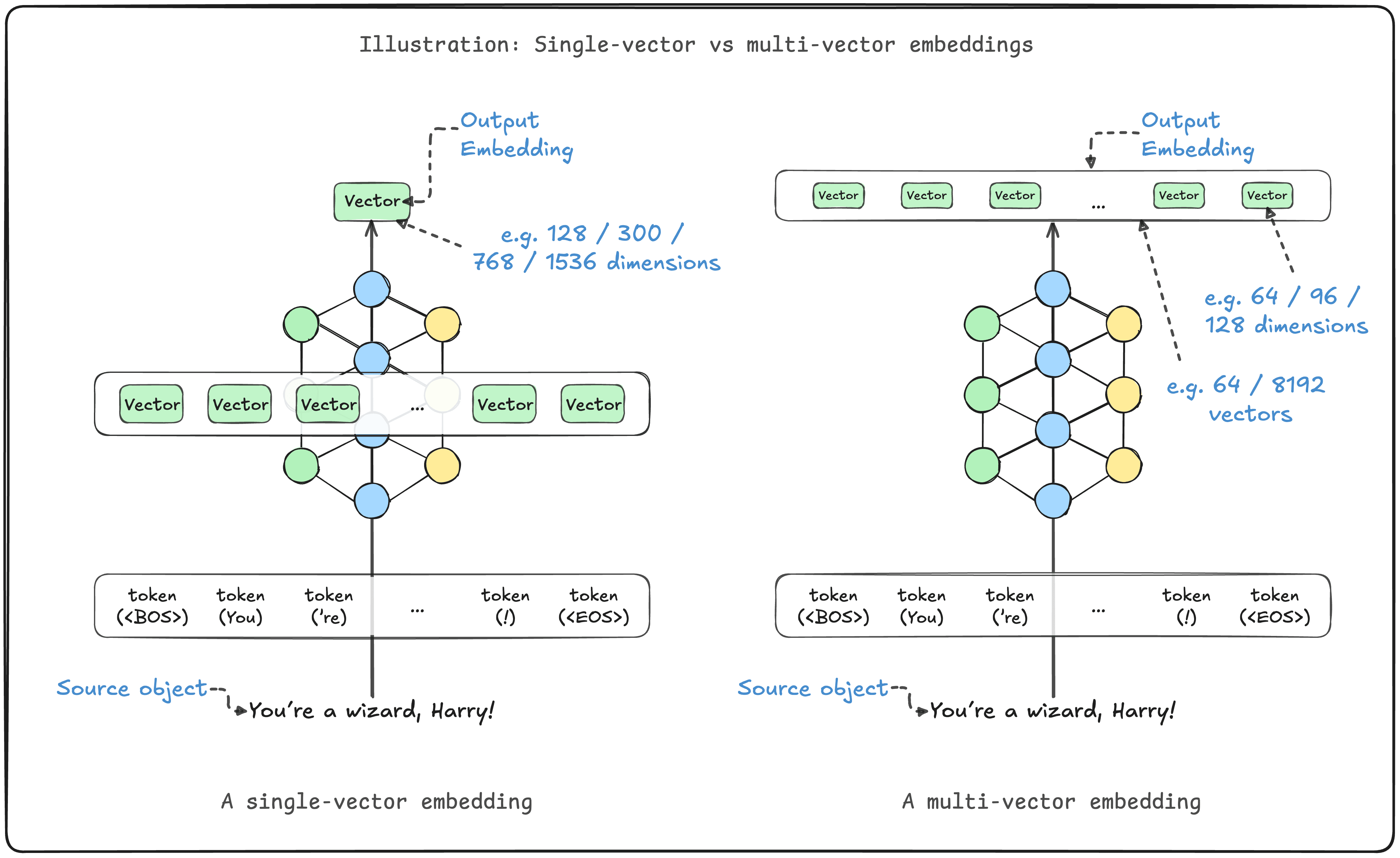
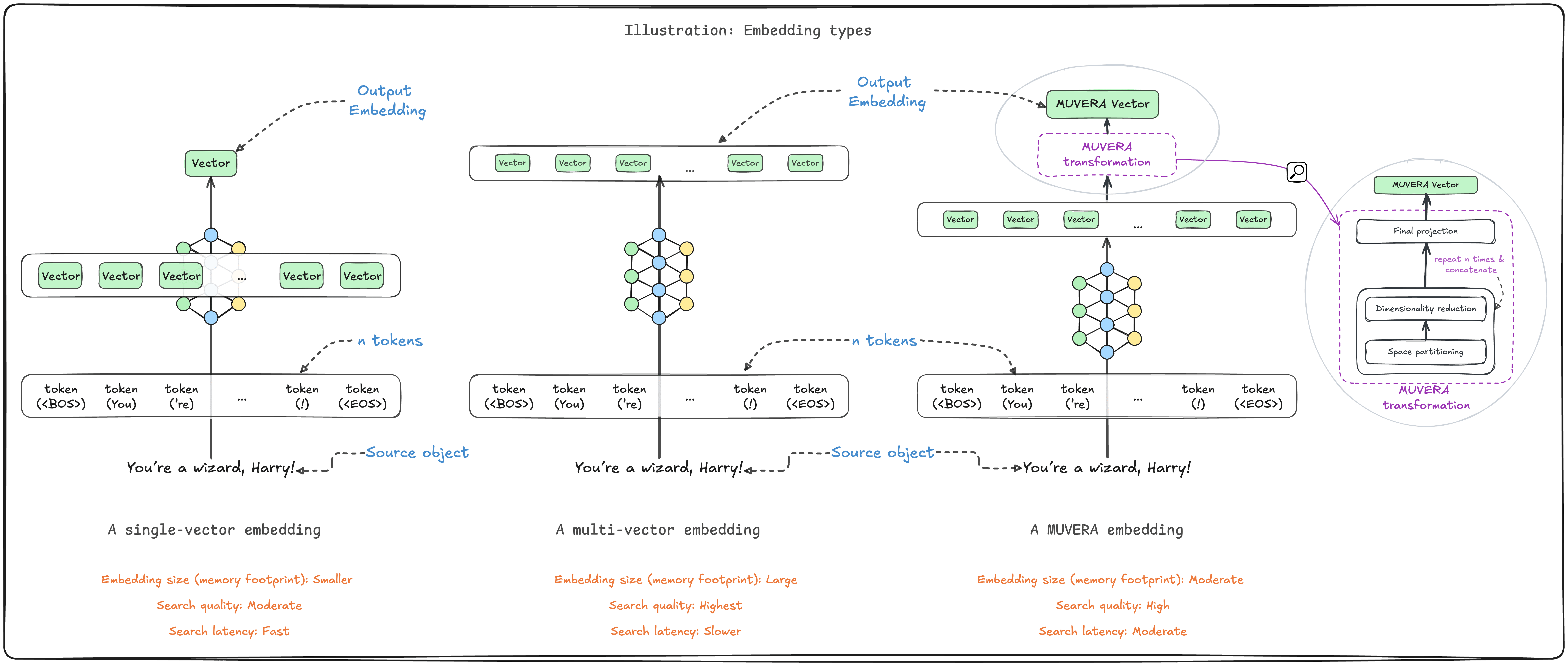
在向量数据库和信息检索领域,多向量嵌入模型(如 ColBERT、ColPali)凭借其强大的语义捕获能力正在成为主流选择。这类模型能够保留文本的词元级别含义,或是识别图像不同部分的信息特征。然而,它们也带来了显著的性能挑战:庞大的内存占用和较慢的检索速度。Weaviate 在 1.31 版本中引入的 MUVERA 编码算法,正是为了解决这些问题而生。
多向量模型的优势与困境
多向量嵌入的核心优势在于其细粒度的语义表达能力。相比单向量模型将整个文档压缩成一个固定长度的向量,多向量模型为文档的每个词元或图像块生成独立的向量表示。这种设计使得模型能够捕捉更丰富的语义信息,在检索任务中展现出更高的准确性。
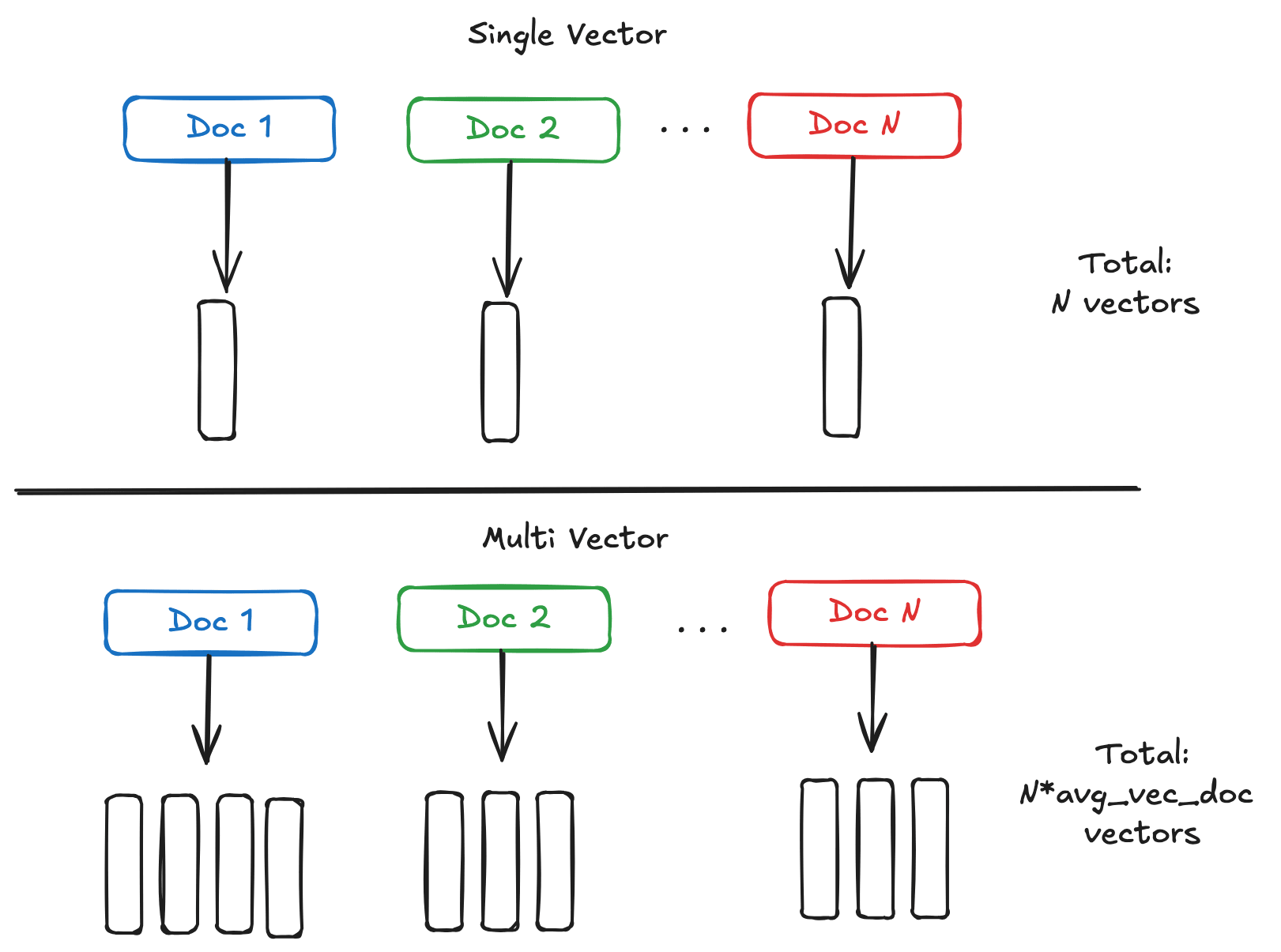
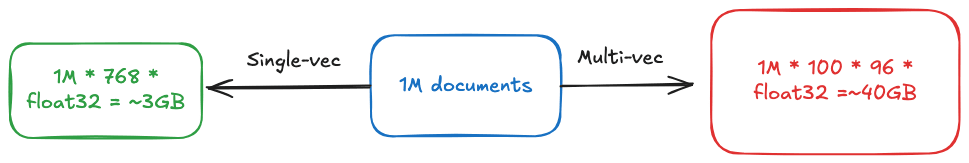
但这种精细化表示的代价同样明显。假设要索引一百万个文档,每个文档平均包含 100 个词元。使用传统的单向量模型(768 维,32 位浮点数),大约需要 3.1GB 内存。而多向量模型(96 维)的内存消耗可能高达 40GB,超过十倍的差距。这种内存压力在大规模部署场景下会转化为实实在在的成本负担。
性能瓶颈不仅体现在存储层面。在检索阶段,多向量模型需要使用 MaxSim 运算符计算相似度。这个过程需要遍历查询的每个词元,找出它与文档所有词元中的最佳匹配,然后累加所有匹配得分。数学表达式如下:
sim(D,Q)=∑q∈Qmaxd∈Dq⋅dsim(D,Q)=\sum_{q \in Q} \max_{d \in D} q \cdot dsim(D,Q)=q∈Q∑d∈Dmaxq⋅d
这种非线性计算相比简单的点积运算复杂得多,直接影响了查询响应速度和数据导入效率。
MUVERA 的核心思想
MUVERA(Multi-Vector Retrieval via Fixed Dimensional Encodings)的设计哲学是将复杂的多向量检索问题转化为单向量最大内积搜索。算法的关键在于构建固定维度编码(FDE),将一组长度不定的向量集合压缩成单个固定长度的向量表示。
整个转换过程可以用一个简洁的映射函数表示:
encode(xmulti)⟹xsinglex∈{D,Q}encode(x_{multi}) \implies x_{single} \quad \quad x \in \{D, Q\}encode(xmulti)⟹xsinglex∈{D,Q}
这里的核心目标是让编码后的单向量点积能够很好地近似原始多向量的 MaxSim 相似度:
maxSim(D,Q)≈dsingle⋅qsinglemaxSim(D,Q) \approx d_{single} \cdot q_{single}maxSim(D,Q)≈dsingle⋅qsingle
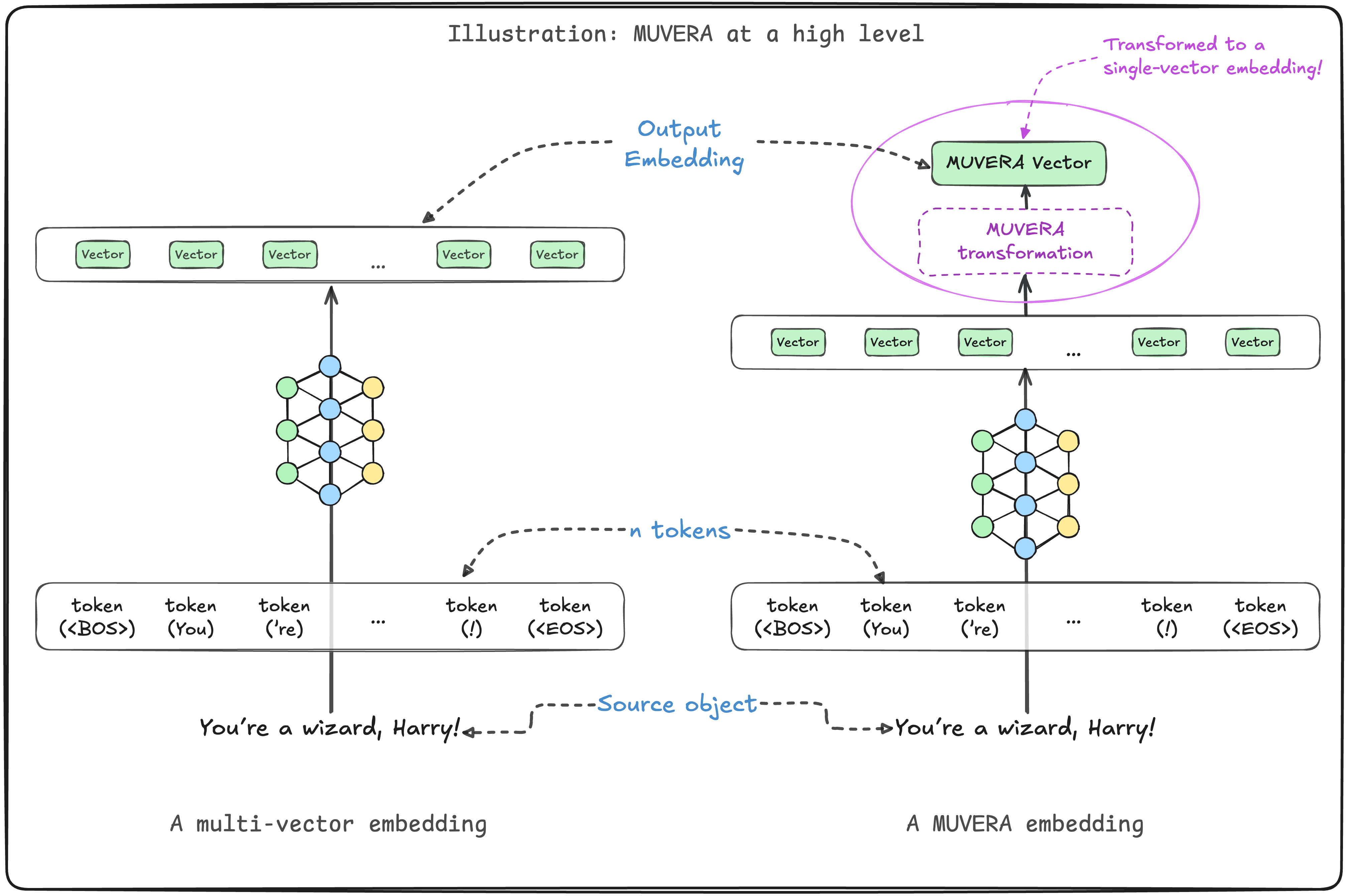
这种转换带来的效率提升是显著的。对于包含 100 万个文档、每个文档 100 个向量的数据集,传统方案需要索引 1 亿个向量,而 MUVERA 只需处理 100 万个 FDE 向量,将 HNSW 图的规模缩减到原来的 1%。
算法实现细节
MUVERA 通过四个精心设计的步骤完成编码转换:空间划分、降维、重复增强和最终投影。每个步骤都有明确的数学基础和实际考量。
空间划分策略
第一步是将高维向量空间划分成若干个桶。算法采用 SimHash 技术实现这一过程,这是一种基于局部敏感哈希的方法。具体来说,算法会采样 ksimk_{sim}ksim 个高斯向量,然后通过计算输入向量与这些高斯向量的点积符号来确定桶编号:
φ(x)=(1(g1⋅x),…,1(gksim⋅x))\varphi(x)=\left(1(g_1 \cdot x), \dots,1(g_{k_{sim}}\cdot x)\right)φ(x)=(1(g1⋅x),…,1(gksim⋅x))
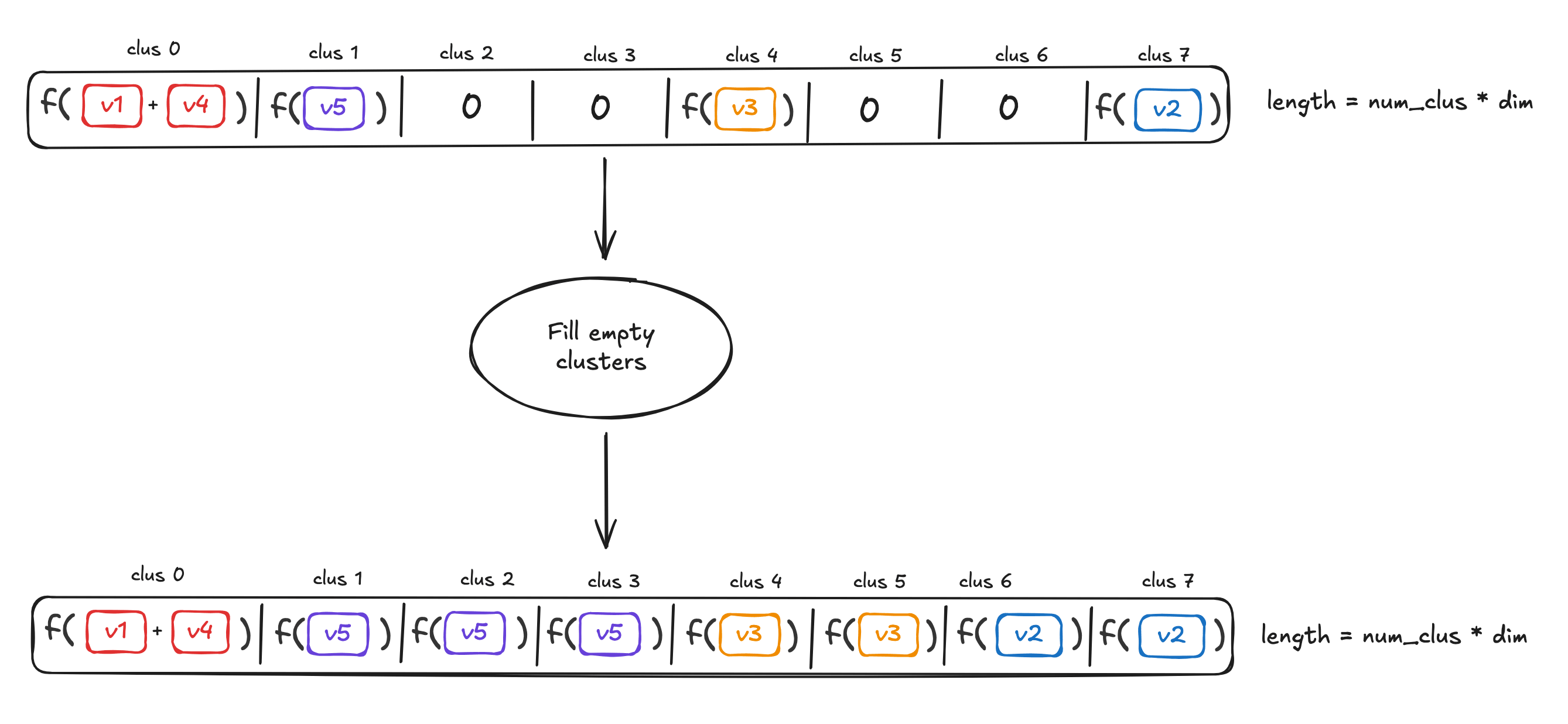
这种划分方式的优势在于其与数据分布无关,不需要预先训练,也不会因为数据漂移而失效。划分完成后,属于同一个桶的向量会被聚合成一个代表性向量。
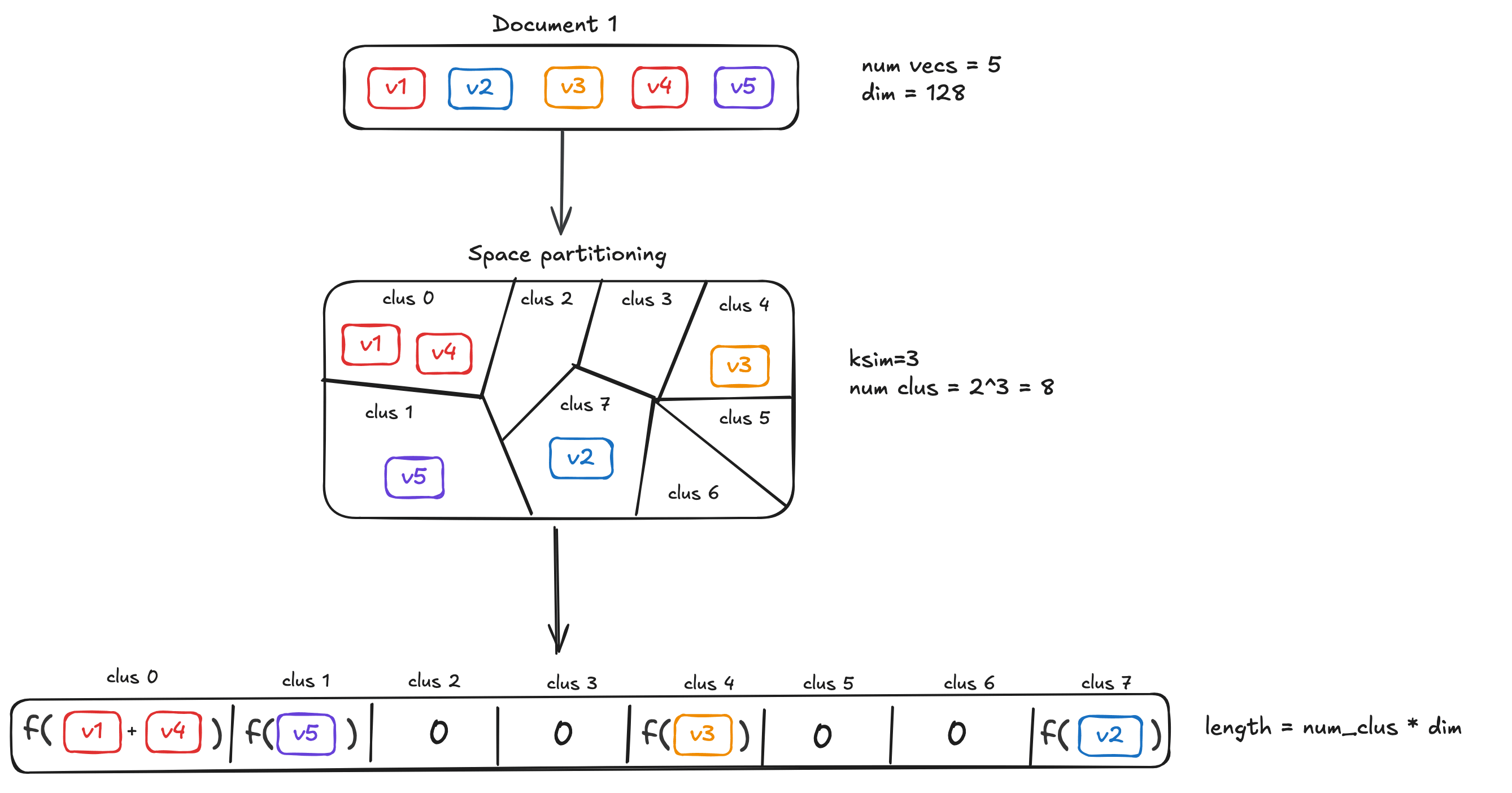
对于文档编码,每个桶的子向量计算方式为:
dk=1∥D∩φ−1(k)∥∑d∈D,φ(d)=kdd_k = \frac{1}{\|D \cap \varphi^{-1}(k)\|} \sum_{d \in D, \varphi(d)=k}ddk=∥D∩φ−1(k)∥1d∈D,φ(d)=k∑d
而查询编码则直接对属于同一桶的向量求和:
qk=∑q∈Q,φ(q)=kqq_k = \sum_{q \in Q, \varphi(q)=k}qqk=q∈Q,φ(q)=k∑q
这种不对称处理恰好对应了 MaxSim 运算的特性。
降维与重复
空间划分后得到的向量维度是 B×dimB \times dimB×dim,其中 BBB 是桶的数量,dimdimdim 是原始向量维度。为了进一步压缩表示,MUVERA 使用随机投影矩阵进行降维:
ψ(dk)=1dproj⋅S⋅dk\psi(d_k)=\frac{1}{\sqrt{d_{proj}}} \cdot S \cdot d_kψ(dk)=dproj1⋅S⋅dk
这里的随机矩阵 SSS 元素取值为 ±1\pm1±1,遵循 Johnson-Lindenstrauss 引理,能够在降维的同时保持向量间点积的相对关系。
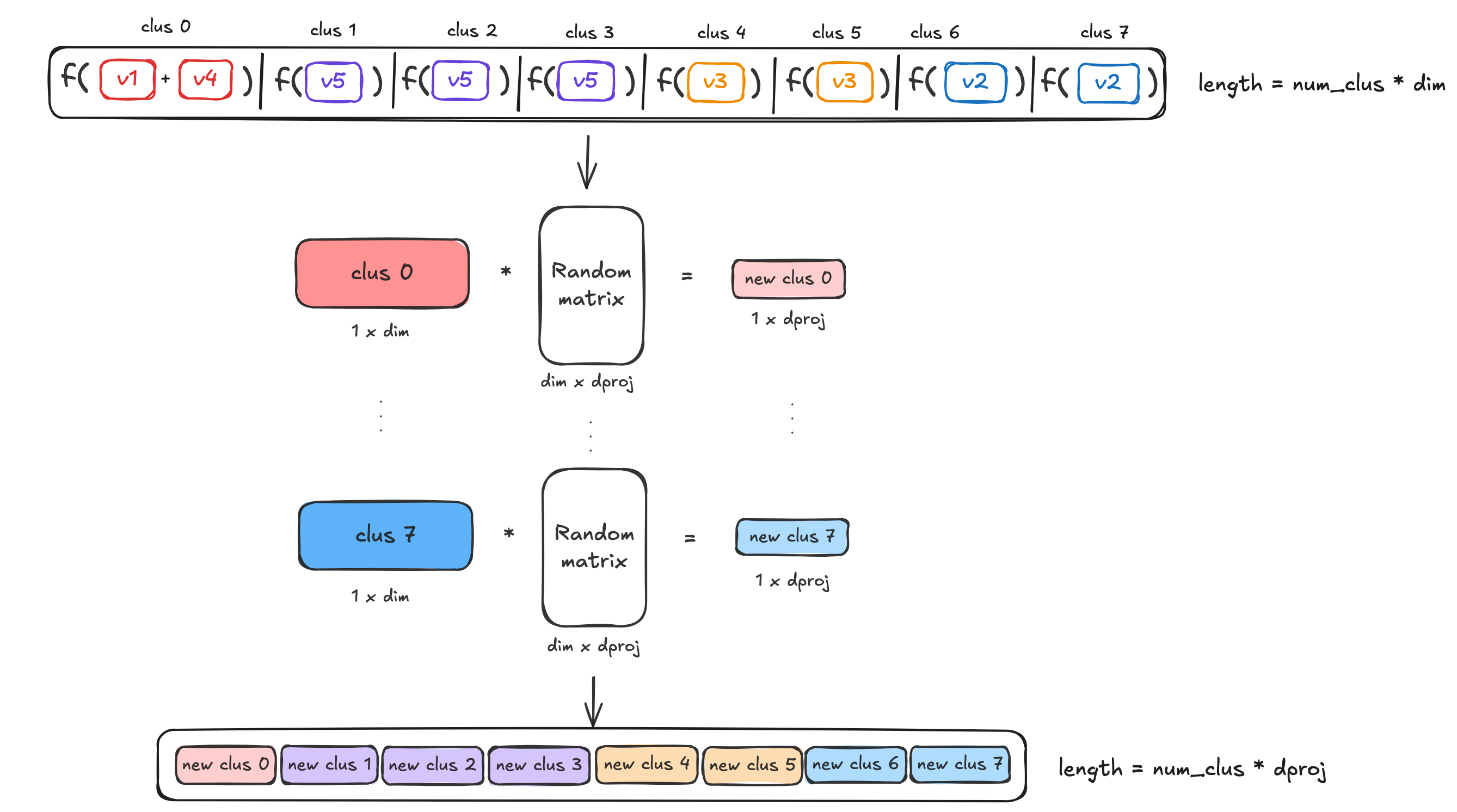
为了提高编码的鲁棒性,算法会重复执行空间划分和降维步骤 RrepsR_{reps}Rreps 次,将得到的多个编码向量拼接起来。最终的 FDE 维度为 Rreps×B×dprojR_{reps} \times B \times d_{proj}Rreps×B×dproj。
性能评测与实际效果
Weaviate 团队使用 LoTTE 基准测试数据集进行了详细的性能评估。该数据集包含约 11.9 万个文档,使用 ColBERT v2.0 编码后生成了 1500 万个 128 维向量,总内存占用约 8GB。
启用 MUVERA 后(参数设置为 ksim=4k_{sim}=4ksim=4,dproj=16d_{proj}=16dproj=16,Rreps=10R_{reps}=10Rreps=10),每个文档被编码为 2560 维的单一向量。这使得总浮点数存储量从 19 亿降至 3.04 亿,内存节省接近 80%。更重要的是,HNSW 图的节点数从 1500 万降至 11.9 万,这对于图遍历效率的提升是质的飞跃。
数据导入速度的改善同样显著。基准场景下,导入 11 万个对象需要 20 多分钟,相当于每秒只能处理约 100 个对象。而使用 MUVERA 后,这个时间缩短到 3-6 分钟。对于需要频繁更新索引的生产环境,这种效率提升意义重大。
性能权衡考量
技术方案从来不是完美的,MUVERA 也有其代价。最主要的妥协体现在召回率上。测试数据显示,在相同的搜索参数下,启用 MUVERA 会导致召回率下降。不过,这个问题可以通过调整 HNSW 的 ef 参数来缓解。
当 ef 值设置在 512 以上时,召回率可以恢复到 80% 以上;而在 2048 时甚至能超过 90%。但提高 ef 值意味着要检索更多的候选集,这会降低查询吞吐量。因此,实际应用中需要在召回质量和查询速度之间找到平衡点。

Google Research 团队的实验结果进一步验证了 MUVERA 的效果。在 BEIR 基准测试中,相比基于单向量启发式的 PLAID 系统,MUVERA 在召回率平均提升 10% 的同时,将延迟降低了 90%。这种性能提升在大规模部署中的价值不言而喻。
适用场景分析
MUVERA 并非万能方案,它最适合以下几类应用场景。首先是内存成本敏感的大规模部署。当数据集规模达到千万甚至亿级时,内存占用的降低可以直接转化为每年数万甚至数十万美元的成本节约。其次是对索引速度有较高要求的场景,比如需要频繁更新的实时系统。
另一个重要考量是对召回质量的容忍度。如果应用场景对检索精度有极致要求,那么需要仔细权衡 MUVERA 带来的召回率下降是否可以接受。不过对于许多实际应用来说,轻微的召回损失往往是可以承受的,特别是考虑到可以通过调整搜索参数来部分恢复性能。
从实现角度看,Weaviate 的集成使得启用 MUVERA 变得非常简单,只需要几行配置代码。用户可以设置的主要参数包括 k_sim(空间划分的细粒度)、d_proj(降维后的维度)和 r_reps(重复次数)。Weaviate 团队为这些参数提供了合理的默认值,大多数场景下可以直接使用。
值得注意的是,MUVERA 的固定维度编码还可以结合标量量化(Scalar Quantization)等技术进一步压缩。Google 的研究表明,通过乘积量化可以在几乎不影响检索质量的前提下,将内存占用再减少 32 倍。这为超大规模应用提供了更多优化空间。
实现
https://github.com/sionic-ai/muvera-py/tree/master
我在github上面找到一个MUVERA的python实现,大家可以尝试一下
import logging
import timeimport numpy as np
from dataclasses import dataclass, replace
from enum import Enum
from typing import Optional, Listclass EncodingType(Enum):DEFAULT_SUM = 0AVERAGE = 1class ProjectionType(Enum):DEFAULT_IDENTITY = 0AMS_SKETCH = 1@dataclass
class FixedDimensionalEncodingConfig:dimension: int = 128num_repetitions: int = 10num_simhash_projections: int = 6seed: int = 42encoding_type: EncodingType = EncodingType.DEFAULT_SUMprojection_type: ProjectionType = ProjectionType.DEFAULT_IDENTITYprojection_dimension: Optional[int] = Nonefill_empty_partitions: bool = Falsefinal_projection_dimension: Optional[int] = Nonedef _append_to_gray_code(gray_code: int, bit: bool) -> int:return (gray_code << 1) + (int(bit) ^ (gray_code & 1))def _gray_code_to_binary(num: int) -> int:mask = num >> 1while mask != 0:num = num ^ maskmask >>= 1return numdef _simhash_matrix_from_seed(dimension: int, num_projections: int, seed: int
) -> np.ndarray:rng = np.random.default_rng(seed)return rng.normal(loc=0.0, scale=1.0, size=(dimension, num_projections)).astype(np.float32)def _ams_projection_matrix_from_seed(dimension: int, projection_dim: int, seed: int
) -> np.ndarray:rng = np.random.default_rng(seed)out = np.zeros((dimension, projection_dim), dtype=np.float32)indices = rng.integers(0, projection_dim, size=dimension)signs = rng.choice([-1.0, 1.0], size=dimension)out[np.arange(dimension), indices] = signsreturn outdef _apply_count_sketch_to_vector(input_vector: np.ndarray, final_dimension: int, seed: int
) -> np.ndarray:rng = np.random.default_rng(seed)out = np.zeros(final_dimension, dtype=np.float32)indices = rng.integers(0, final_dimension, size=input_vector.shape[0])signs = rng.choice([-1.0, 1.0], size=input_vector.shape[0])np.add.at(out, indices, signs * input_vector)return outdef _simhash_partition_index_gray(sketch_vector: np.ndarray) -> int:partition_index = 0for val in sketch_vector:partition_index = _append_to_gray_code(partition_index, val > 0)return partition_indexdef _distance_to_simhash_partition(sketch_vector: np.ndarray, partition_index: int
) -> int:num_projections = sketch_vector.sizebinary_representation = _gray_code_to_binary(partition_index)sketch_bits = (sketch_vector > 0).astype(int)binary_array = (binary_representation >> np.arange(num_projections - 1, -1, -1)) & 1return int(np.sum(sketch_bits != binary_array))def _generate_fde_internal(point_cloud: np.ndarray, config: FixedDimensionalEncodingConfig
) -> np.ndarray:if point_cloud.ndim != 2 or point_cloud.shape[1] != config.dimension:raise ValueError(f"Input data shape {point_cloud.shape} is inconsistent with config dimension {config.dimension}.")if not (0 <= config.num_simhash_projections < 32):raise ValueError(f"num_simhash_projections must be in [0, 31]: {config.num_simhash_projections}")num_points, original_dim = point_cloud.shapenum_partitions = 2**config.num_simhash_projectionsuse_identity_proj = config.projection_type == ProjectionType.DEFAULT_IDENTITYprojection_dim = original_dim if use_identity_proj else config.projection_dimensionif not use_identity_proj and (not projection_dim or projection_dim <= 0):raise ValueError("A positive projection_dimension is required for non-identity projections.")final_fde_dim = config.num_repetitions * num_partitions * projection_dimout_fde = np.zeros(final_fde_dim, dtype=np.float32)for rep_num in range(config.num_repetitions):current_seed = config.seed + rep_numsketches = point_cloud @ _simhash_matrix_from_seed(original_dim, config.num_simhash_projections, current_seed)if use_identity_proj:projected_matrix = point_cloudelif config.projection_type == ProjectionType.AMS_SKETCH:ams_matrix = _ams_projection_matrix_from_seed(original_dim, projection_dim, current_seed)projected_matrix = point_cloud @ ams_matrixrep_fde_sum = np.zeros(num_partitions * projection_dim, dtype=np.float32)partition_counts = np.zeros(num_partitions, dtype=np.int32)partition_indices = np.array([_simhash_partition_index_gray(sketches[i]) for i in range(num_points)])for i in range(num_points):start_idx = partition_indices[i] * projection_dimrep_fde_sum[start_idx : start_idx + projection_dim] += projected_matrix[i]partition_counts[partition_indices[i]] += 1if config.encoding_type == EncodingType.AVERAGE:for i in range(num_partitions):start_idx = i * projection_dimif partition_counts[i] > 0:rep_fde_sum[start_idx : start_idx + projection_dim] /= (partition_counts[i])elif config.fill_empty_partitions and num_points > 0:distances = [_distance_to_simhash_partition(sketches[j], i)for j in range(num_points)]nearest_point_idx = np.argmin(distances)rep_fde_sum[start_idx : start_idx + projection_dim] = (projected_matrix[nearest_point_idx])rep_start_index = rep_num * num_partitions * projection_dimout_fde[rep_start_index : rep_start_index + rep_fde_sum.size] = rep_fde_sumif config.final_projection_dimension and config.final_projection_dimension > 0:return _apply_count_sketch_to_vector(out_fde, config.final_projection_dimension, config.seed)return out_fdedef generate_query_fde(point_cloud: np.ndarray, config: FixedDimensionalEncodingConfig
) -> np.ndarray:"""Generates a Fixed Dimensional Encoding for a query point cloud (using SUM)."""if config.fill_empty_partitions:raise ValueError("Query FDE generation does not support 'fill_empty_partitions'.")query_config = replace(config, encoding_type=EncodingType.DEFAULT_SUM)return _generate_fde_internal(point_cloud, query_config)def generate_document_fde(point_cloud: np.ndarray, config: FixedDimensionalEncodingConfig
) -> np.ndarray:"""Generates a Fixed Dimensional Encoding for a document point cloud (using AVERAGE)."""doc_config = replace(config, encoding_type=EncodingType.AVERAGE)return _generate_fde_internal(point_cloud, doc_config)def generate_fde(point_cloud: np.ndarray, config: FixedDimensionalEncodingConfig
) -> np.ndarray:if config.encoding_type == EncodingType.DEFAULT_SUM:return generate_query_fde(point_cloud, config)elif config.encoding_type == EncodingType.AVERAGE:return generate_document_fde(point_cloud, config)else:raise ValueError(f"Unsupported encoding type in config: {config.encoding_type}")def generate_document_fde_batch(doc_embeddings_list: List[np.ndarray], config: FixedDimensionalEncodingConfig
) -> np.ndarray:"""Generates FDEs for a batch of documents using highly optimized NumPy vectorization.Fully compliant with C++ implementation including all projection types."""batch_start_time = time.perf_counter()num_docs = len(doc_embeddings_list)if num_docs == 0:logging.warning("[FDE Batch] Empty document list provided")return np.array([])logging.info(f"[FDE Batch] Starting batch FDE generation for {num_docs} documents")# Input validationvalid_docs = []for i, doc in enumerate(doc_embeddings_list):if doc.ndim != 2:logging.warning(f"[FDE Batch] Document {i} has invalid shape (ndim={doc.ndim}), skipping")continueif doc.shape[1] != config.dimension:raise ValueError(f"Document {i} has incorrect dimension: expected {config.dimension}, got {doc.shape[1]}")if doc.shape[0] == 0:logging.warning(f"[FDE Batch] Document {i} has no vectors, skipping")continuevalid_docs.append(doc)if len(valid_docs) == 0:logging.warning("[FDE Batch] No valid documents after filtering")return np.array([])num_docs = len(valid_docs)doc_embeddings_list = valid_docs# Determine projection dimension (matching C++ logic)use_identity_proj = config.projection_type == ProjectionType.DEFAULT_IDENTITYif use_identity_proj:projection_dim = config.dimensionlogging.info(f"[FDE Batch] Using identity projection (dim={projection_dim})")else:if not config.projection_dimension or config.projection_dimension <= 0:raise ValueError("A positive projection_dimension must be specified for non-identity projections")projection_dim = config.projection_dimensionlogging.info(f"[FDE Batch] Using {config.projection_type.name} projection: "f"{config.dimension} -> {projection_dim}")# Configuration summarynum_partitions = 2**config.num_simhash_projectionslogging.info(f"[FDE Batch] Configuration: {config.num_repetitions} repetitions, "f"{num_partitions} partitions, projection_dim={projection_dim}")# Document trackingdoc_lengths = np.array([len(doc) for doc in doc_embeddings_list], dtype=np.int32)total_vectors = np.sum(doc_lengths)doc_boundaries = np.insert(np.cumsum(doc_lengths), 0, 0)doc_indices = np.repeat(np.arange(num_docs), doc_lengths)logging.info(f"[FDE Batch] Total vectors: {total_vectors}, avg per doc: {total_vectors / num_docs:.1f}")# Concatenate all embeddingsconcat_start = time.perf_counter()all_points = np.vstack(doc_embeddings_list).astype(np.float32)concat_time = time.perf_counter() - concat_startlogging.info(f"[FDE Batch] Concatenation completed in {concat_time:.3f}s")# Pre-allocate outputfinal_fde_dim = config.num_repetitions * num_partitions * projection_dimout_fdes = np.zeros((num_docs, final_fde_dim), dtype=np.float32)logging.info(f"[FDE Batch] Output FDE dimension: {final_fde_dim}")# Process each repetitionfor rep_num in range(config.num_repetitions):# rep_start_time = time.perf_counter()current_seed = config.seed + rep_numif rep_num % 5 == 0: # Log every 5 repetitionslogging.info(f"[FDE Batch] Processing repetition {rep_num + 1}/{config.num_repetitions}")# Step 1: SimHash projectionsimhash_start = time.perf_counter()simhash_matrix = _simhash_matrix_from_seed(config.dimension, config.num_simhash_projections, current_seed)all_sketches = all_points @ simhash_matrixsimhash_time = time.perf_counter() - simhash_start# Step 2: Apply dimensionality reduction if configuredproj_start = time.perf_counter()if use_identity_proj:projected_points = all_pointselif config.projection_type == ProjectionType.AMS_SKETCH:ams_matrix = _ams_projection_matrix_from_seed(config.dimension, projection_dim, current_seed)projected_points = all_points @ ams_matrixelse:raise ValueError(f"Unsupported projection type: {config.projection_type}")proj_time = time.perf_counter() - proj_start# Step 3: Vectorized partition index calculationpartition_start = time.perf_counter()bits = (all_sketches > 0).astype(np.uint32)partition_indices = np.zeros(total_vectors, dtype=np.uint32)# Vectorized Gray Code computationfor bit_idx in range(config.num_simhash_projections):partition_indices = (partition_indices << 1) + (bits[:, bit_idx] ^ (partition_indices & 1))partition_time = time.perf_counter() - partition_start# Step 4: Vectorized aggregationagg_start = time.perf_counter()# Initialize storage for this repetitionrep_fde_sum = np.zeros((num_docs * num_partitions * projection_dim,), dtype=np.float32)partition_counts = np.zeros((num_docs, num_partitions), dtype=np.int32)# Count vectors per partition per documentnp.add.at(partition_counts, (doc_indices, partition_indices), 1)# Aggregate vectors using flattened indexing for efficiencydoc_part_indices = doc_indices * num_partitions + partition_indicesbase_indices = doc_part_indices * projection_dimfor d in range(projection_dim):flat_indices = base_indices + dnp.add.at(rep_fde_sum, flat_indices, projected_points[:, d])# Reshape for easier manipulationrep_fde_sum = rep_fde_sum.reshape(num_docs, num_partitions, projection_dim)agg_time = time.perf_counter() - agg_start# Step 5: Convert sums to averages (for document FDE)avg_start = time.perf_counter()# Vectorized division where counts > 0non_zero_mask = partition_counts > 0counts_3d = partition_counts[:, :, np.newaxis] # Broadcasting for division# Safe division (avoid divide by zero)np.divide(rep_fde_sum, counts_3d, out=rep_fde_sum, where=counts_3d > 0)# Fill empty partitions if configuredempty_filled = 0if config.fill_empty_partitions:empty_mask = ~non_zero_maskempty_docs, empty_parts = np.where(empty_mask)for doc_idx, part_idx in zip(empty_docs, empty_parts):if doc_lengths[doc_idx] == 0:continue# Get sketches for this documentdoc_start = doc_boundaries[doc_idx]doc_end = doc_boundaries[doc_idx + 1]doc_sketches = all_sketches[doc_start:doc_end]# Vectorized distance calculationbinary_rep = _gray_code_to_binary(part_idx)target_bits = (binary_rep >> np.arange(config.num_simhash_projections - 1, -1, -1)) & 1distances = np.sum((doc_sketches > 0).astype(int) != target_bits, axis=1)nearest_local_idx = np.argmin(distances)nearest_global_idx = doc_start + nearest_local_idxrep_fde_sum[doc_idx, part_idx, :] = projected_points[nearest_global_idx]empty_filled += 1avg_time = time.perf_counter() - avg_start# Step 6: Copy results to output arrayrep_output_start = rep_num * num_partitions * projection_dimout_fdes[:, rep_output_start : rep_output_start + num_partitions * projection_dim] = rep_fde_sum.reshape(num_docs, -1)# Log timing for first repetitionif rep_num == 0:logging.info("[FDE Batch] Repetition timing breakdown:")logging.info(f" - SimHash: {simhash_time:.3f}s")logging.info(f" - Projection: {proj_time:.3f}s")logging.info(f" - Partition indices: {partition_time:.3f}s")logging.info(f" - Aggregation: {agg_time:.3f}s")logging.info(f" - Averaging: {avg_time:.3f}s")if config.fill_empty_partitions:logging.info(f" - Filled {empty_filled} empty partitions")# Step 7: Apply final projection if configuredif config.final_projection_dimension and config.final_projection_dimension > 0:logging.info(f"[FDE Batch] Applying final projection: {final_fde_dim} -> "f"{config.final_projection_dimension}")final_proj_start = time.perf_counter()# Process in chunks to avoid memory issueschunk_size = min(100, num_docs)final_fdes = []for i in range(0, num_docs, chunk_size):chunk_end = min(i + chunk_size, num_docs)chunk_fdes = np.array([_apply_count_sketch_to_vector(out_fdes[j], config.final_projection_dimension, config.seed)for j in range(i, chunk_end)])final_fdes.append(chunk_fdes)out_fdes = np.vstack(final_fdes)final_proj_time = time.perf_counter() - final_proj_startlogging.info(f"[FDE Batch] Final projection completed in {final_proj_time:.3f}s")# Final statistics and validationtotal_time = time.perf_counter() - batch_start_timelogging.info(f"[FDE Batch] Batch generation completed in {total_time:.3f}s")logging.info(f"[FDE Batch] Average time per document: {total_time / num_docs * 1000:.2f}ms")logging.info(f"[FDE Batch] Throughput: {num_docs / total_time:.1f} docs/sec")logging.info(f"[FDE Batch] Output shape: {out_fdes.shape}")# Validate output dimensionsexpected_dim = (final_fde_dimif not config.final_projection_dimensionelse config.final_projection_dimension)assert out_fdes.shape == (num_docs, expected_dim), (f"Output shape mismatch: {out_fdes.shape} != ({num_docs}, {expected_dim})")# doc_config = replace(config, encoding_type=EncodingType.AVERAGE)return out_fdesif __name__ == "__main__":print(f"\n{'=' * 20} SCENARIO 1: Basic FDE Generation {'=' * 20}")base_config = FixedDimensionalEncodingConfig(dimension=128, num_repetitions=2, num_simhash_projections=4, seed=42)query_data = np.random.randn(32, base_config.dimension).astype(np.float32)doc_data = np.random.randn(80, base_config.dimension).astype(np.float32)query_fde = generate_query_fde(query_data, base_config)doc_fde = generate_document_fde(doc_data, replace(base_config, fill_empty_partitions=True))expected_dim = (base_config.num_repetitions* (2**base_config.num_simhash_projections)* base_config.dimension)print(f"Query FDE Shape: {query_fde.shape} (Expected: {expected_dim})")print(f"Document FDE Shape: {doc_fde.shape} (Expected: {expected_dim})")print(f"Similarity Score: {np.dot(query_fde, doc_fde):.4f}")assert query_fde.shape[0] == expected_dimprint(f"\n{'=' * 20} SCENARIO 2: Inner Projection (AMS Sketch) {'=' * 20}")ams_config = replace(base_config, projection_type=ProjectionType.AMS_SKETCH, projection_dimension=16)query_fde_ams = generate_query_fde(query_data, ams_config)expected_dim_ams = (ams_config.num_repetitions* (2**ams_config.num_simhash_projections)* ams_config.projection_dimension)print(f"AMS Sketch FDE Shape: {query_fde_ams.shape} (Expected: {expected_dim_ams})")assert query_fde_ams.shape[0] == expected_dim_amsprint(f"\n{'=' * 20} SCENARIO 3: Final Projection (Count Sketch) {'=' * 20}")final_proj_config = replace(base_config, final_projection_dimension=1024)query_fde_final = generate_query_fde(query_data, final_proj_config)print(f"Final Projection FDE Shape: {query_fde_final.shape} (Expected: {final_proj_config.final_projection_dimension})")assert query_fde_final.shape[0] == final_proj_config.final_projection_dimensionprint(f"\n{'=' * 20} SCENARIO 4: Top-level `generate_fde` wrapper {'=' * 20}")query_fde_2 = generate_fde(query_data, replace(base_config, encoding_type=EncodingType.DEFAULT_SUM))doc_fde_2 = generate_fde(doc_data, replace(base_config, encoding_type=EncodingType.AVERAGE))print(f"Wrapper-generated Query FDE is identical: {np.allclose(query_fde, query_fde_2)}")print(f"Wrapper-generated Document FDE is identical: {np.allclose(doc_fde, doc_fde_2)}")print("\nAll test scenarios completed successfully.")
结语
随着 ColBERT、ColPali 等多向量模型的进一步发展,以及 MUVERA 这类优化算法的不断演进,多向量检索的效率瓶颈正在逐步被克服。未来,在推荐系统、搜索引擎、文档检索等场景中,多向量技术很可能成为标准配置。而 MUVERA 所展示的将复杂问题简化为经典问题的思路,也为其他领域的算法优化提供了有价值的参考。
