批次量生成不同方向形变结构脚本
这是一个用于晶体结构分析和应变模拟的Python工具,可以读取VASP格式的POSCAR文件,分析晶体系统类型,并生成不同应变条件下的新POSCAR文件。
运行环境要求:
numpy>=1.20.0
matplotlib>=3.5.0
tqdm>=4.60.0
生成的应变结构文件会保存在指定的输出目录中(默认为"strained_structures"),文件名包含了应变类型和参数信息。
执行程序方法,将文章后附的代码写进一个UTF-8格式的文本文件中,重命名为dis-latt_optimized.py
执行
python dis-latt_optimized.py选择POSCAR文件
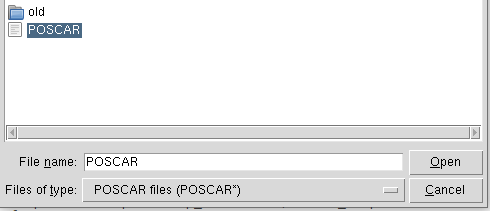
POSCAR信息提示
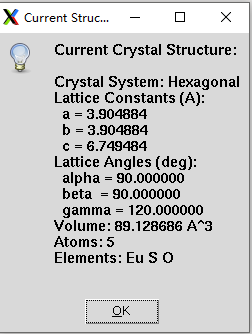
程序界面
首次运行可能需要强行关闭tk的空白窗口
施加x方向应变并确认
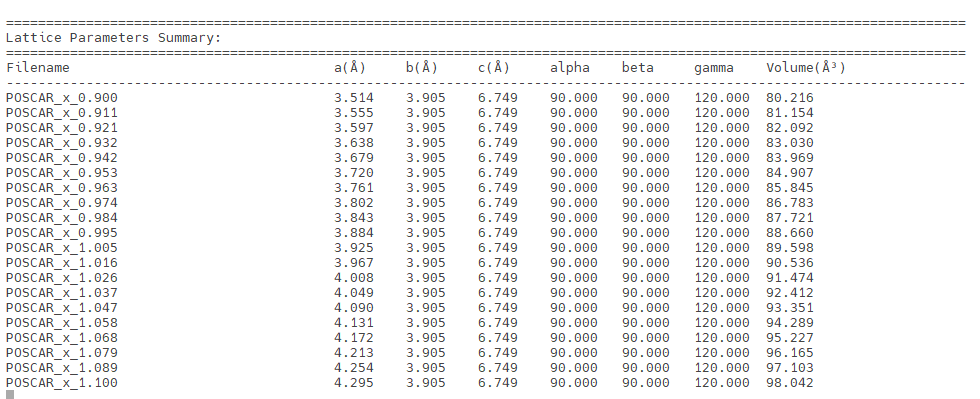
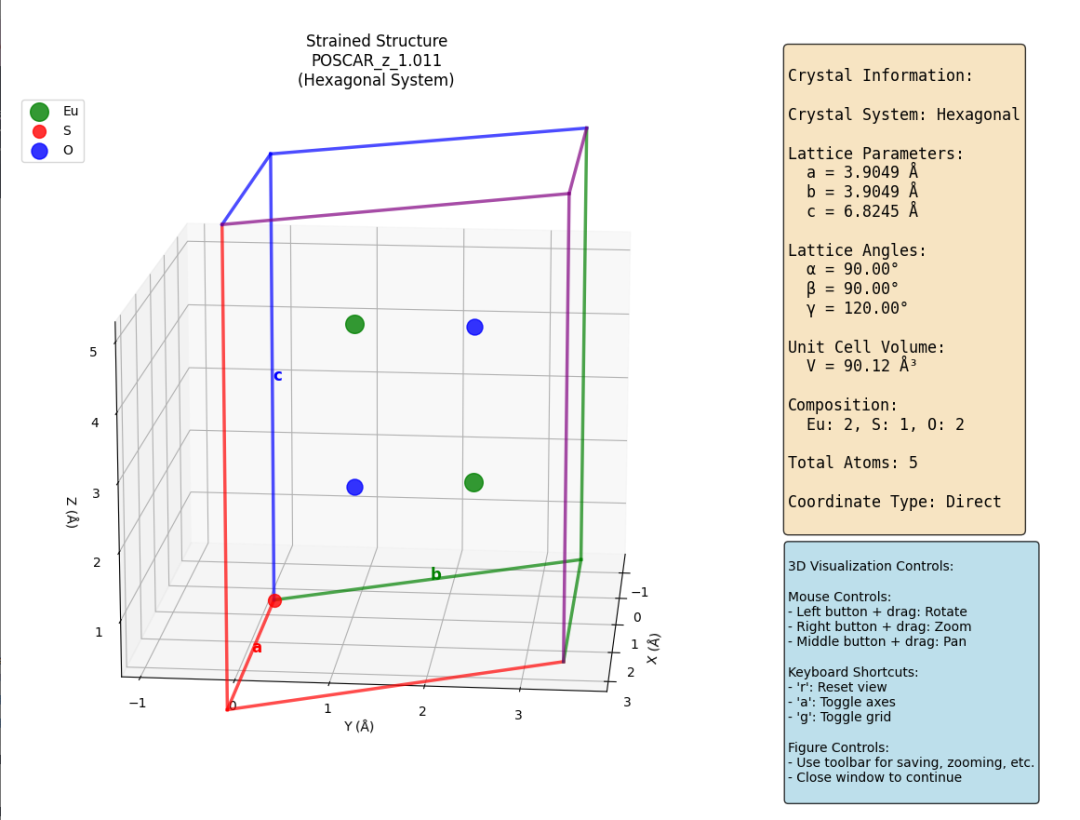
生成文件的简易可视化
点击continue的yes即可继续进行文件生成
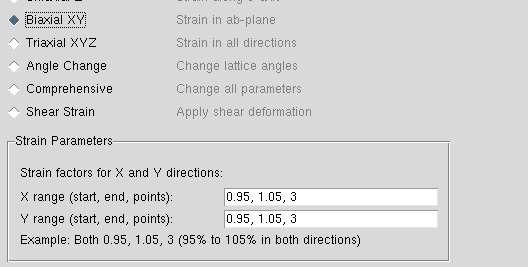
xy的批次量应变生成
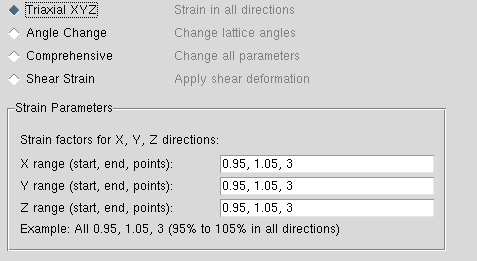
xyz的批次量应变生成
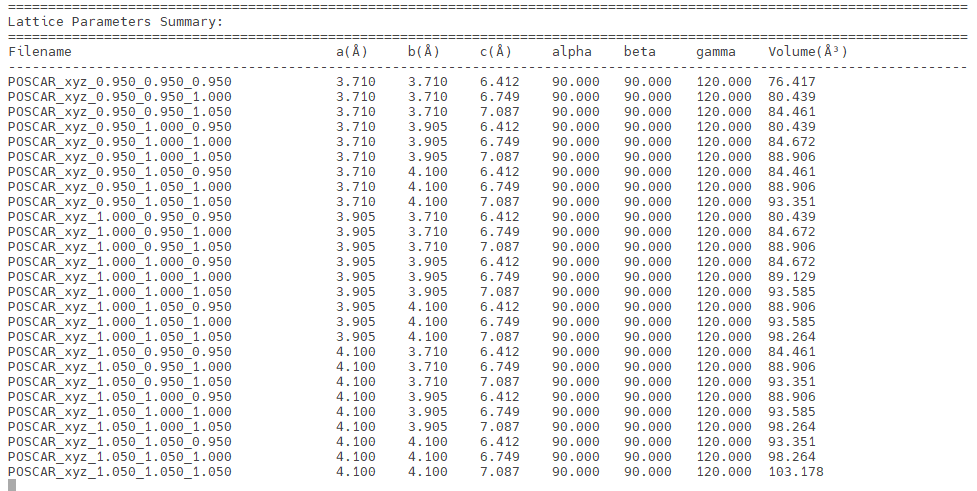
最后点击continue的no即可结束程序
完整代码
#!/usr/bin/env python3
# -*- coding: utf-8 -*-import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import os
import math
import sys# Set matplotlib backend
import matplotlib
matplotlib.use('TkAgg')if sys.platform.startswith('linux'):matplotlib.rcParams['font.family'] = 'DejaVu Sans'try:import tkinter as tkfrom tkinter import ttk, messagebox, simpledialog, filedialog
except ImportError:print("Tkinter is not available. Please install it to use the GUI features.")sys.exit(1)class InteractiveCrystalStrain:def __init__(self, poscar_file=None):self.poscar_file = poscar_fileif poscar_file and os.path.exists(poscar_file):self.read_poscar()self.analyze_crystal_system()self.calculate_lattice_parameters()else:self.ask_for_poscar()def ask_for_poscar(self):"""Ask for POSCAR file path"""root = tk.Tk()root.withdraw()self.poscar_file = filedialog.askopenfilename(title="Select POSCAR file",filetypes=[("POSCAR files", "POSCAR*"), ("All files", "*.*")])if not self.poscar_file:messagebox.showerror("Error", "No POSCAR file selected. Program will exit.")sys.exit(1)self.read_poscar()self.analyze_crystal_system()self.calculate_lattice_parameters()root.destroy()def read_poscar(self):"""Read POSCAR file"""with open(self.poscar_file, 'r') as f:lines = f.readlines()self.title = lines[0].strip()self.scale_factor = float(lines[1].strip())self.lattice_vectors = []for i in range(2, 5):vector = [float(x) for x in lines[i].split()]self.lattice_vectors.append(vector)self.lattice_vectors = np.array(self.lattice_vectors)self.elements = lines[5].split()self.element_counts = [int(x) for x in lines[6].split()]self.total_atoms = sum(self.element_counts)self.coord_type = lines[7].strip().lower()self.atomic_coords = []for i in range(8, 8 + self.total_atoms):coords = [float(x) for x in lines[i].split()[:3]]self.atomic_coords.append(coords)self.atomic_coords = np.array(self.atomic_coords)def analyze_crystal_system(self):"""Analyze crystal system"""a1, a2, a3 = self.lattice_vectorsa = np.linalg.norm(a1)b = np.linalg.norm(a2)c = np.linalg.norm(a3)alpha = np.degrees(np.arccos(np.dot(a2, a3) / (b * c)))beta = np.degrees(np.arccos(np.dot(a1, a3) / (a * c)))gamma = np.degrees(np.arccos(np.dot(a1, a2) / (a * b)))tol = 1e-3if (abs(a - b) < tol and abs(b - c) < tol and abs(alpha - 90) < tol and abs(beta - 90) < tol and abs(gamma - 90) < tol):self.crystal_system = "Cubic"elif (abs(a - b) < tol and abs(b - c) < tol and abs(alpha - beta) < tol and abs(beta - gamma) < tol and abs(alpha - 90) > tol):self.crystal_system = "Rhombohedral"elif (abs(a - b) < tol and abs(b - c) > tol and abs(alpha - 90) < tol and abs(beta - 90) < tol and abs(gamma - 90) < tol):self.crystal_system = "Tetragonal"elif (abs(a - b) < tol and abs(b - c) > tol and abs(alpha - 90) < tol and abs(beta - 90) < tol and abs(gamma - 120) < tol):self.crystal_system = "Hexagonal"elif (abs(a - b) > tol and abs(b - c) > tol and abs(a - c) > tol and abs(alpha - 90) < tol and abs(beta - 90) < tol and abs(gamma - 90) < tol):self.crystal_system = "Orthorhombic"elif (abs(alpha - 90) > tol and abs(beta - 90) < tol and abs(gamma - 90) < tol):self.crystal_system = "Monoclinic"else:self.crystal_system = "Triclinic"def calculate_lattice_parameters(self):"""Calculate lattice parameters"""a1, a2, a3 = self.lattice_vectorsself.a = np.linalg.norm(a1)self.b = np.linalg.norm(a2)self.c = np.linalg.norm(a3)self.alpha = np.degrees(np.arccos(np.dot(a2, a3) / (self.b * self.c)))self.beta = np.degrees(np.arccos(np.dot(a1, a3) / (self.a * self.c)))self.gamma = np.degrees(np.arccos(np.dot(a1, a2) / (self.a * self.b)))self.volume = np.abs(np.dot(a1, np.cross(a2, a3)))def show_current_parameters(self):"""Show current lattice parameters"""message = f"""Current Crystal Structure:Crystal System: {self.crystal_system}
Lattice Constants (A):a = {self.a:.6f}b = {self.b:.6f}c = {self.c:.6f}
Lattice Angles (deg):alpha = {self.alpha:.6f}beta = {self.beta:.6f}gamma = {self.gamma:.6f}
Volume: {self.volume:.6f} A^3
Atoms: {self.total_atoms}
Elements: {' '.join(self.elements)}
"""messagebox.showinfo("Current Structure", message)def get_user_input(self):"""Get detailed user input for strain parameters"""root = tk.Tk()root.title("Strain Parameters")root.geometry("700x600")# Make sure the window is on toproot.lift()root.attributes('-topmost', True)root.after_idle(root.attributes, '-topmost', False)main_frame = ttk.Frame(root, padding="10")main_frame.grid(row=0, column=0, sticky=(tk.W, tk.E, tk.N, tk.S))# Strain type selectionttk.Label(main_frame, text="Select Strain Type:", font=("Arial", 10, "bold")).grid(row=0, column=0, columnspan=2, pady=10, sticky=tk.W)strain_types = [("Uniaxial X", "x", "Strain along a-axis"),("Uniaxial Y", "y", "Strain along b-axis"), ("Uniaxial Z", "z", "Strain along c-axis"),("Biaxial XY", "xy", "Strain in ab-plane"),("Triaxial XYZ", "xyz", "Strain in all directions"),("Angle Change", "angle", "Change lattice angles"),("Comprehensive", "all", "Change all parameters"),("Shear Strain", "shear", "Apply shear deformation")]self.strain_type = tk.StringVar(value="x")for i, (text, value, desc) in enumerate(strain_types):ttk.Radiobutton(main_frame, text=text, variable=self.strain_type, value=value).grid(row=1+i, column=0, sticky=tk.W, pady=2)ttk.Label(main_frame, text=desc, foreground="gray").grid(row=1+i, column=1, sticky=tk.W, pady=2)# Parameter input based on strain typeparam_frame = ttk.LabelFrame(main_frame, text="Strain Parameters", padding="10")param_frame.grid(row=len(strain_types)+2, column=0, columnspan=2, sticky=(tk.W, tk.E), pady=10)# Dynamic parameter fieldsself.param_entries = {}def update_parameter_fields():# Clear previous fieldsfor widget in param_frame.winfo_children():widget.destroy()strain_type = self.strain_type.get()row = 0if strain_type in ['x', 'y', 'z']:ttk.Label(param_frame, text=f"Strain factor for {strain_type.upper()} direction:").grid(row=row, column=0, sticky=tk.W, pady=5)ttk.Label(param_frame, text="Range (start, end, points):").grid(row=row+1, column=0, sticky=tk.W, pady=2)self.param_entries['range'] = ttk.Entry(param_frame, width=30)self.param_entries['range'].grid(row=row+1, column=1, pady=2)self.param_entries['range'].insert(0, "0.9, 1.1, 5")ttk.Label(param_frame, text="Example: 0.9, 1.1, 5 (90% to 110% compression to stretch, 5 points)").grid(row=row+2, column=0, columnspan=2, sticky=tk.W, pady=2)elif strain_type == 'xy':ttk.Label(param_frame, text="Strain factors for X and Y directions:").grid(row=row, column=0, sticky=tk.W, pady=5)ttk.Label(param_frame, text="X range (start, end, points):").grid(row=row+1, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="Y range (start, end, points):").grid(row=row+2, column=0, sticky=tk.W, pady=2)self.param_entries['x_range'] = ttk.Entry(param_frame, width=30)self.param_entries['x_range'].grid(row=row+1, column=1, pady=2)self.param_entries['x_range'].insert(0, "0.95, 1.05, 3")self.param_entries['y_range'] = ttk.Entry(param_frame, width=30)self.param_entries['y_range'].grid(row=row+2, column=1, pady=2)self.param_entries['y_range'].insert(0, "0.95, 1.05, 3")ttk.Label(param_frame, text="Example: Both 0.95, 1.05, 3 (95% to 105% in both directions)").grid(row=row+3, column=0, columnspan=2, sticky=tk.W, pady=2)elif strain_type == 'xyz':ttk.Label(param_frame, text="Strain factors for X, Y, Z directions:").grid(row=row, column=0, sticky=tk.W, pady=5)ttk.Label(param_frame, text="X range (start, end, points):").grid(row=row+1, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="Y range (start, end, points):").grid(row=row+2, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="Z range (start, end, points):").grid(row=row+3, column=0, sticky=tk.W, pady=2)self.param_entries['x_range'] = ttk.Entry(param_frame, width=30)self.param_entries['x_range'].grid(row=row+1, column=1, pady=2)self.param_entries['x_range'].insert(0, "0.95, 1.05, 3")self.param_entries['y_range'] = ttk.Entry(param_frame, width=30)self.param_entries['y_range'].grid(row=row+2, column=1, pady=2)self.param_entries['y_range'].insert(0, "0.95, 1.05, 3")self.param_entries['z_range'] = ttk.Entry(param_frame, width=30)self.param_entries['z_range'].grid(row=row+3, column=1, pady=2)self.param_entries['z_range'].insert(0, "0.95, 1.05, 3")ttk.Label(param_frame, text="Example: All 0.95, 1.05, 3 (95% to 105% in all directions)").grid(row=row+4, column=0, columnspan=2, sticky=tk.W, pady=2)elif strain_type == 'angle':ttk.Label(param_frame, text="Change lattice angles (degrees):").grid(row=row, column=0, sticky=tk.W, pady=5)ttk.Label(param_frame, text="Alpha range (start, end, points):").grid(row=row+1, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="Beta range (start, end, points):").grid(row=row+2, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="Gamma range (start, end, points):").grid(row=row+3, column=0, sticky=tk.W, pady=2)self.param_entries['alpha_range'] = ttk.Entry(param_frame, width=30)self.param_entries['alpha_range'].grid(row=row+1, column=1, pady=2)self.param_entries['alpha_range'].insert(0, f"{self.alpha}, {self.alpha}, 1")self.param_entries['beta_range'] = ttk.Entry(param_frame, width=30)self.param_entries['beta_range'].grid(row=row+2, column=1, pady=2)self.param_entries['beta_range'].insert(0, f"{self.beta}, {self.beta}, 1")self.param_entries['gamma_range'] = ttk.Entry(param_frame, width=30)self.param_entries['gamma_range'].grid(row=row+3, column=1, pady=2)self.param_entries['gamma_range'].insert(0, f"{self.gamma-5}, {self.gamma+5}, 5")ttk.Label(param_frame, text=f"Current angles: alpha={self.alpha:.2f}, beta={self.beta:.2f}, gamma={self.gamma:.2f}").grid(row=row+4, column=0, columnspan=2, sticky=tk.W, pady=2)elif strain_type == 'all':ttk.Label(param_frame, text="Comprehensive parameter change:").grid(row=row, column=0, sticky=tk.W, pady=5)ttk.Label(param_frame, text="a scaling (start, end, points):").grid(row=row+1, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="b scaling (start, end, points):").grid(row=row+2, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="c scaling (start, end, points):").grid(row=row+3, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="alpha (start, end, points):").grid(row=row+4, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="beta (start, end, points):").grid(row=row+5, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="gamma (start, end, points):").grid(row=row+6, column=0, sticky=tk.W, pady=2)self.param_entries['a_range'] = ttk.Entry(param_frame, width=30)self.param_entries['a_range'].grid(row=row+1, column=1, pady=2)self.param_entries['a_range'].insert(0, "0.98, 1.02, 3")self.param_entries['b_range'] = ttk.Entry(param_frame, width=30)self.param_entries['b_range'].grid(row=row+2, column=1, pady=2)self.param_entries['b_range'].insert(0, "0.98, 1.02, 3")self.param_entries['c_range'] = ttk.Entry(param_frame, width=30)self.param_entries['c_range'].grid(row=row+3, column=1, pady=2)self.param_entries['c_range'].insert(0, "0.98, 1.02, 3")self.param_entries['alpha_range'] = ttk.Entry(param_frame, width=30)self.param_entries['alpha_range'].grid(row=row+4, column=1, pady=2)self.param_entries['alpha_range'].insert(0, f"{self.alpha}, {self.alpha}, 1")self.param_entries['beta_range'] = ttk.Entry(param_frame, width=30)self.param_entries['beta_range'].grid(row=row+5, column=1, pady=2)self.param_entries['beta_range'].insert(0, f"{self.beta}, {self.beta}, 1")self.param_entries['gamma_range'] = ttk.Entry(param_frame, width=30)self.param_entries['gamma_range'].grid(row=row+6, column=1, pady=2)self.param_entries['gamma_range'].insert(0, f"{self.gamma-2}, {self.gamma+2}, 3")elif strain_type == 'shear':ttk.Label(param_frame, text="Shear strain components:").grid(row=row, column=0, sticky=tk.W, pady=5)ttk.Label(param_frame, text="XY shear (start, end, points):").grid(row=row+1, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="XZ shear (start, end, points):").grid(row=row+2, column=0, sticky=tk.W, pady=2)ttk.Label(param_frame, text="YZ shear (start, end, points):").grid(row=row+3, column=0, sticky=tk.W, pady=2)self.param_entries['xy_range'] = ttk.Entry(param_frame, width=30)self.param_entries['xy_range'].grid(row=row+1, column=1, pady=2)self.param_entries['xy_range'].insert(0, "-0.1, 0.1, 5")self.param_entries['xz_range'] = ttk.Entry(param_frame, width=30)self.param_entries['xz_range'].grid(row=row+2, column=1, pady=2)self.param_entries['xz_range'].insert(0, "0, 0, 1")self.param_entries['yz_range'] = ttk.Entry(param_frame, width=30)self.param_entries['yz_range'].grid(row=row+3, column=1, pady=2)self.param_entries['yz_range'].insert(0, "0, 0, 1")ttk.Label(param_frame, text="Example: XY from -0.1 to 0.1 (shear in xy plane)").grid(row=row+4, column=0, columnspan=2, sticky=tk.W, pady=2)# Initial setupupdate_parameter_fields()# Update fields when strain type changesfor rb in main_frame.winfo_children():if isinstance(rb, ttk.Radiobutton):rb.configure(command=update_parameter_fields)# Output directoryttk.Label(main_frame, text="Output Directory:").grid(row=len(strain_types)+3, column=0, sticky=tk.W, pady=10)self.output_dir = tk.StringVar(value="strained_structures")ttk.Entry(main_frame, textvariable=self.output_dir, width=30).grid(row=len(strain_types)+3, column=1, pady=10)# Advanced optionsadv_frame = ttk.LabelFrame(main_frame, text="Advanced Options", padding="5")adv_frame.grid(row=len(strain_types)+4, column=0, columnspan=2, sticky=(tk.W, tk.E), pady=10)self.preserve_volume = tk.BooleanVar()ttk.Checkbutton(adv_frame, text="Preserve Volume", variable=self.preserve_volume).grid(row=0, column=0, sticky=tk.W)self.preserve_angles = tk.BooleanVar()ttk.Checkbutton(adv_frame, text="Preserve Angles", variable=self.preserve_angles).grid(row=0, column=1, sticky=tk.W)# Buttonsbutton_frame = ttk.Frame(main_frame)button_frame.grid(row=len(strain_types)+5, column=0, columnspan=2, pady=20)self.user_input = Nonedef on_ok():# Collect all parametersself.user_input = {'strain_type': self.strain_type.get(),'output_dir': self.output_dir.get(),'preserve_volume': self.preserve_volume.get(),'preserve_angles': self.preserve_angles.get(),'parameters': {}}# Parse parameter rangesstrain_type = self.strain_type.get()if strain_type in ['x', 'y', 'z']:range_str = self.param_entries['range'].get()try:start, end, points = [float(x.strip()) for x in range_str.split(',')]self.user_input['parameters']['range'] = (start, end, int(points))except:messagebox.showerror("Error", "Invalid range format. Use: start, end, points")returnelif strain_type == 'xy':try:x_range = [float(x.strip()) for x in self.param_entries['x_range'].get().split(',')]y_range = [float(x.strip()) for x in self.param_entries['y_range'].get().split(',')]self.user_input['parameters']['x_range'] = (x_range[0], x_range[1], int(x_range[2]))self.user_input['parameters']['y_range'] = (y_range[0], y_range[1], int(y_range[2]))except:messagebox.showerror("Error", "Invalid range format. Use: start, end, points")returnelif strain_type == 'xyz':try:x_range = [float(x.strip()) for x in self.param_entries['x_range'].get().split(',')]y_range = [float(x.strip()) for x in self.param_entries['y_range'].get().split(',')]z_range = [float(x.strip()) for x in self.param_entries['z_range'].get().split(',')]self.user_input['parameters']['x_range'] = (x_range[0], x_range[1], int(x_range[2]))self.user_input['parameters']['y_range'] = (y_range[0], y_range[1], int(y_range[2]))self.user_input['parameters']['z_range'] = (z_range[0], z_range[1], int(z_range[2]))except:messagebox.showerror("Error", "Invalid range format. Use: start, end, points")returnelif strain_type == 'angle':try:alpha_range = [float(x.strip()) for x in self.param_entries['alpha_range'].get().split(',')]beta_range = [float(x.strip()) for x in self.param_entries['beta_range'].get().split(',')]gamma_range = [float(x.strip()) for x in self.param_entries['gamma_range'].get().split(',')]self.user_input['parameters']['alpha_range'] = (alpha_range[0], alpha_range[1], int(alpha_range[2]))self.user_input['parameters']['beta_range'] = (beta_range[0], beta_range[1], int(beta_range[2]))self.user_input['parameters']['gamma_range'] = (gamma_range[0], gamma_range[1], int(gamma_range[2]))except:messagebox.showerror("Error", "Invalid range format. Use: start, end, points")returnelif strain_type == 'all':try:a_range = [float(x.strip()) for x in self.param_entries['a_range'].get().split(',')]b_range = [float(x.strip()) for x in self.param_entries['b_range'].get().split(',')]c_range = [float(x.strip()) for x in self.param_entries['c_range'].get().split(',')]alpha_range = [float(x.strip()) for x in self.param_entries['alpha_range'].get().split(',')]beta_range = [float(x.strip()) for x in self.param_entries['beta_range'].get().split(',')]gamma_range = [float(x.strip()) for x in self.param_entries['gamma_range'].get().split(',')]self.user_input['parameters']['a_range'] = (a_range[0], a_range[1], int(a_range[2]))self.user_input['parameters']['b_range'] = (b_range[0], b_range[1], int(b_range[2]))self.user_input['parameters']['c_range'] = (c_range[0], c_range[1], int(c_range[2]))self.user_input['parameters']['alpha_range'] = (alpha_range[0], alpha_range[1], int(alpha_range[2]))self.user_input['parameters']['beta_range'] = (beta_range[0], beta_range[1], int(beta_range[2]))self.user_input['parameters']['gamma_range'] = (gamma_range[0], gamma_range[1], int(gamma_range[2]))except:messagebox.showerror("Error", "Invalid range format. Use: start, end, points")returnelif strain_type == 'shear':try:xy_range = [float(x.strip()) for x in self.param_entries['xy_range'].get().split(',')]xz_range = [float(x.strip()) for x in self.param_entries['xz_range'].get().split(',')]yz_range = [float(x.strip()) for x in self.param_entries['yz_range'].get().split(',')]self.user_input['parameters']['xy_range'] = (xy_range[0], xy_range[1], int(xy_range[2]))self.user_input['parameters']['xz_range'] = (xz_range[0], xz_range[1], int(xz_range[2]))self.user_input['parameters']['yz_range'] = (yz_range[0], yz_range[1], int(yz_range[2]))except:messagebox.showerror("Error", "Invalid range format. Use: start, end, points")returnroot.destroy()def on_cancel():self.user_input = Noneroot.destroy()ttk.Button(button_frame, text="OK", command=on_ok).grid(row=0, column=0, padx=5)ttk.Button(button_frame, text="Cancel", command=on_cancel).grid(row=0, column=1, padx=5)root.mainloop()return self.user_inputdef apply_strain(self, strain_type, strain_values, output_dir="strained_structures", preserve_angles=False, preserve_volume=False):"""Apply strain to crystal structure"""if not os.path.exists(output_dir):os.makedirs(output_dir)results = []for strain_value in strain_values:strained_lattice = self.lattice_vectors.copy()strained_coords = self.atomic_coords.copy()if strain_type == 'x':strained_lattice[0] *= strain_valueif preserve_volume:factor = 1.0 / math.sqrt(strain_value)strained_lattice[1] *= factorstrained_lattice[2] *= factorelif strain_type == 'y':strained_lattice[1] *= strain_valueif preserve_volume:factor = 1.0 / math.sqrt(strain_value)strained_lattice[0] *= factorstrained_lattice[2] *= factorelif strain_type == 'z':strained_lattice[2] *= strain_valueif preserve_volume:factor = 1.0 / math.sqrt(strain_value)strained_lattice[0] *= factorstrained_lattice[1] *= factorelif strain_type == 'xy':if isinstance(strain_value, (list, tuple)) and len(strain_value) == 2:strained_lattice[0] *= strain_value[0]strained_lattice[1] *= strain_value[1]else:strained_lattice[0] *= strain_valuestrained_lattice[1] *= strain_valueif preserve_volume:factor = 1.0 / (strain_value[0] * strain_value[1]) if isinstance(strain_value, (list, tuple)) else 1.0 / strain_valuestrained_lattice[2] *= factorelif strain_type == 'xyz':if isinstance(strain_value, (list, tuple)) and len(strain_value) == 3:strained_lattice[0] *= strain_value[0]strained_lattice[1] *= strain_value[1]strained_lattice[2] *= strain_value[2]else:strained_lattice *= strain_valueelif strain_type == 'angle':if isinstance(strain_value, (list, tuple)) and len(strain_value) == 3:new_alpha, new_beta, new_gamma = strain_valueelse:new_alpha, new_beta, new_gamma = self.alpha, self.beta, strain_valuestrained_lattice = self.build_lattice_from_parameters(self.a, self.b, self.c, new_alpha, new_beta, new_gamma)elif strain_type == 'all':a_factor, b_factor, c_factor, new_alpha, new_beta, new_gamma = strain_valuenew_a = self.a * a_factornew_b = self.b * b_factornew_c = self.c * c_factorstrained_lattice = self.build_lattice_from_parameters(new_a, new_b, new_c, new_alpha, new_beta, new_gamma)elif strain_type == 'shear':if isinstance(strain_value, (list, tuple)) and len(strain_value) == 3:shear_xy, shear_xz, shear_yz = strain_valueshear_matrix = np.array([[1, shear_xy, shear_xz],[0, 1, shear_yz],[0, 0, 1]])strained_lattice = np.dot(shear_matrix, strained_lattice.T).T# If preserving angles is requestedif preserve_angles and strain_type not in ['angle', 'all']:new_a = np.linalg.norm(strained_lattice[0])new_b = np.linalg.norm(strained_lattice[1])new_c = np.linalg.norm(strained_lattice[2])strained_lattice = self.build_lattice_from_parameters(new_a, new_b, new_c, self.alpha, self.beta, self.gamma)# Calculate new lattice parametersnew_params = self.calculate_lattice_parameters_from_vectors(strained_lattice)# Generate output filenamefilename = self.generate_filename(strain_type, strain_value)output_file = os.path.join(output_dir, filename)# Write new POSCAR fileself.write_poscar(strained_lattice, strained_coords, output_file)results.append({'file': output_file,'strain_type': strain_type,'strain_value': strain_value,'lattice_vectors': strained_lattice,**new_params})return resultsdef build_lattice_from_parameters(self, a, b, c, alpha, beta, gamma):"""Build lattice vectors from lattice parameters"""alpha_rad = np.radians(alpha)beta_rad = np.radians(beta)gamma_rad = np.radians(gamma)a1 = np.array([a, 0, 0])a2 = np.array([b * np.cos(gamma_rad), b * np.sin(gamma_rad), 0])a3_x = c * np.cos(beta_rad)a3_y = c * (np.cos(alpha_rad) - np.cos(beta_rad) * np.cos(gamma_rad)) / np.sin(gamma_rad)a3_z = np.sqrt(c**2 - a3_x**2 - a3_y**2)a3 = np.array([a3_x, a3_y, a3_z])return np.array([a1, a2, a3])def calculate_lattice_parameters_from_vectors(self, lattice_vectors):"""Calculate lattice parameters from lattice vectors"""a1, a2, a3 = lattice_vectorsa = np.linalg.norm(a1)b = np.linalg.norm(a2)c = np.linalg.norm(a3)alpha = np.degrees(np.arccos(np.dot(a2, a3) / (b * c)))beta = np.degrees(np.arccos(np.dot(a1, a3) / (a * c)))gamma = np.degrees(np.arccos(np.dot(a1, a2) / (a * b)))volume = np.abs(np.dot(a1, np.cross(a2, a3)))return {'a': a, 'b': b, 'c': c,'alpha': alpha, 'beta': beta, 'gamma': gamma,'volume': volume}def generate_filename(self, strain_type, strain_value):"""Generate output filename"""if strain_type in ['x', 'y', 'z']:return f"POSCAR_{strain_type}_{strain_value:.3f}"elif strain_type == 'xy':if isinstance(strain_value, (list, tuple)):return f"POSCAR_xy_{strain_value[0]:.3f}_{strain_value[1]:.3f}"else:return f"POSCAR_xy_{strain_value:.3f}"elif strain_type == 'xyz':if isinstance(strain_value, (list, tuple)):return f"POSCAR_xyz_{strain_value[0]:.3f}_{strain_value[1]:.3f}_{strain_value[2]:.3f}"else:return f"POSCAR_xyz_{strain_value:.3f}"elif strain_type == 'angle':if isinstance(strain_value, (list, tuple)):return f"POSCAR_angle_{strain_value[0]:.3f}_{strain_value[1]:.3f}_{strain_value[2]:.3f}"else:return f"POSCAR_angle_{strain_value:.3f}"elif strain_type == 'all':return f"POSCAR_all_{'_'.join(f'{v:.3f}' for v in strain_value)}"elif strain_type == 'shear':return f"POSCAR_shear_{'_'.join(f'{v:.3f}' for v in strain_value)}"else:return f"POSCAR_modified"def write_poscar(self, lattice_vectors, atomic_coords, output_file):"""Write new POSCAR file"""with open(output_file, 'w') as f:f.write(f"{self.title}_modified\n")f.write(" 1.00000000000000\n")for vector in lattice_vectors:f.write(" {:.15f} {:.15f} {:.15f}\n".format(*vector))f.write(" " + " ".join(self.elements) + "\n")f.write(" " + " ".join(map(str, self.element_counts)) + "\n")f.write("Direct\n")for coords in atomic_coords:f.write(" {:.15f} {:.15f} {:.15f}\n".format(*coords))def generate_strain_series(self, user_input):"""Generate a series of strained structures based on user input"""strain_type = user_input['strain_type']params = user_input['parameters']output_dir = user_input['output_dir']preserve_volume = user_input['preserve_volume']preserve_angles = user_input['preserve_angles']strain_values = []if strain_type in ['x', 'y', 'z']:start, end, points = params['range']strain_values = np.linspace(start, end, points)elif strain_type == 'xy':x_start, x_end, x_points = params['x_range']y_start, y_end, y_points = params['y_range']x_values = np.linspace(x_start, x_end, x_points)y_values = np.linspace(y_start, y_end, y_points)# Create all combinationsfor x in x_values:for y in y_values:strain_values.append((x, y))elif strain_type == 'xyz':x_start, x_end, x_points = params['x_range']y_start, y_end, y_points = params['y_range']z_start, z_end, z_points = params['z_range']x_values = np.linspace(x_start, x_end, x_points)y_values = np.linspace(y_start, y_end, y_points)z_values = np.linspace(z_start, z_end, z_points)# Create all combinationsfor x in x_values:for y in y_values:for z in z_values:strain_values.append((x, y, z))elif strain_type == 'angle':alpha_start, alpha_end, alpha_points = params['alpha_range']beta_start, beta_end, beta_points = params['beta_range']gamma_start, gamma_end, gamma_points = params['gamma_range']alpha_values = np.linspace(alpha_start, alpha_end, alpha_points)beta_values = np.linspace(beta_start, beta_end, beta_points)gamma_values = np.linspace(gamma_start, gamma_end, gamma_points)# Create all combinationsfor alpha in alpha_values:for beta in beta_values:for gamma in gamma_values:strain_values.append((alpha, beta, gamma))elif strain_type == 'all':a_start, a_end, a_points = params['a_range']b_start, b_end, b_points = params['b_range']c_start, c_end, c_points = params['c_range']alpha_start, alpha_end, alpha_points = params['alpha_range']beta_start, beta_end, beta_points = params['beta_range']gamma_start, gamma_end, gamma_points = params['gamma_range']a_values = np.linspace(a_start, a_end, a_points)b_values = np.linspace(b_start, b_end, b_points)c_values = np.linspace(c_start, c_end, c_points)alpha_values = np.linspace(alpha_start, alpha_end, alpha_points)beta_values = np.linspace(beta_start, beta_end, beta_points)gamma_values = np.linspace(gamma_start, gamma_end, gamma_points)# Create all combinations (this can get large!)# We'll limit to a reasonable number or use a different approach# For now, let's just take the first few combinationscount = 0max_combinations = 27 # Limit to prevent too many filesfor a in a_values:for b in b_values:for c in c_values:for alpha in alpha_values:for beta in beta_values:for gamma in gamma_values:if count < max_combinations:strain_values.append((a, b, c, alpha, beta, gamma))count += 1else:breakif count >= max_combinations:breakif count >= max_combinations:breakif count >= max_combinations:breakif count >= max_combinations:breakif count >= max_combinations:breakelif strain_type == 'shear':xy_start, xy_end, xy_points = params['xy_range']xz_start, xz_end, xz_points = params['xz_range']yz_start, yz_end, yz_points = params['yz_range']xy_values = np.linspace(xy_start, xy_end, xy_points)xz_values = np.linspace(xz_start, xz_end, xz_points)yz_values = np.linspace(yz_start, yz_end, yz_points)# Create all combinationsfor xy in xy_values:for xz in xz_values:for yz in yz_values:strain_values.append((xy, xz, yz))return self.apply_strain(strain_type, strain_values, output_dir, preserve_angles, preserve_volume)def plot_structure_3d(self, lattice_vectors=None, atomic_coords=None, title="Crystal Structure"):"""Enhanced 3D visualization of crystal structure with crystal info panel"""if lattice_vectors is None:lattice_vectors = self.lattice_vectorsif atomic_coords is None:atomic_coords = self.atomic_coords# Create a figure with two subplots: one for 3D visualization, one for crystal infofig = plt.figure(figsize=(16, 10))# 3D subplotax = fig.add_subplot(121, projection='3d')# Text info subplotax_info = fig.add_subplot(122)ax_info.axis('off') # Hide axes for info panel# Plot the crystal structurecartesian_coords = np.dot(atomic_coords, lattice_vectors)# Plot unit cellself.plot_unit_cell(ax, lattice_vectors)# Plot atomselement_symbols = []for element, count in zip(self.elements, self.element_counts):element_symbols.extend([element] * count)unique_elements = list(set(element_symbols))colors_atoms = ['red', 'blue', 'green', 'orange', 'purple', 'brown', 'pink', 'gray']color_map = {elem: colors_atoms[i % len(colors_atoms)] for i, elem in enumerate(unique_elements)}size_map = {elem: 100 + i * 50 for i, elem in enumerate(unique_elements)}plotted_elements = set()for i, (coord, elem) in enumerate(zip(cartesian_coords, element_symbols)):label = elem if elem not in plotted_elements else ""ax.scatter(*coord, color=color_map[elem], s=size_map[elem], label=label, alpha=0.8)plotted_elements.add(elem)# Set 3D plot propertiesax.set_xlabel('X (Å)')ax.set_ylabel('Y (Å)')ax.set_zlabel('Z (Å)')ax.set_title(f"{title}\n({self.crystal_system} System)")# Add legendax.legend(loc='upper left', bbox_to_anchor=(0, 1))# Set equal aspect ratiomax_range = max(np.ptp(cartesian_coords, axis=0))mid_x = np.mean(cartesian_coords[:, 0])mid_y = np.mean(cartesian_coords[:, 1])mid_z = np.mean(cartesian_coords[:, 2])ax.set_xlim(mid_x - max_range/2, mid_x + max_range/2)ax.set_ylim(mid_y - max_range/2, mid_y + max_range/2)ax.set_zlim(mid_z - max_range/2, mid_z + max_range/2)# Calculate current lattice parameters for displaycurrent_params = self.calculate_lattice_parameters_from_vectors(lattice_vectors)# Create crystal information textinfo_text = f"""
Crystal Information:Crystal System: {self.crystal_system}Lattice Parameters:a = {current_params['a']:.4f} Åb = {current_params['b']:.4f} Åc = {current_params['c']:.4f} ÅLattice Angles:α = {current_params['alpha']:.2f}°β = {current_params['beta']:.2f}°γ = {current_params['gamma']:.2f}°Unit Cell Volume:V = {current_params['volume']:.2f} ųComposition:{', '.join([f'{elem}: {count}' for elem, count in zip(self.elements, self.element_counts)])}Total Atoms: {self.total_atoms}Coordinate Type: {self.coord_type.title()}
"""# Add crystal information to the info panelax_info.text(0.05, 0.95, info_text, transform=ax_info.transAxes, fontsize=12,verticalalignment='top', horizontalalignment='left',bbox=dict(boxstyle='round', facecolor='wheat', alpha=0.8),fontfamily='monospace')# Add instructions for interactioninstructions = """
3D Visualization Controls:Mouse Controls:
- Left button + drag: Rotate
- Right button + drag: Zoom
- Middle button + drag: PanKeyboard Shortcuts:
- 'r': Reset view
- 'a': Toggle axes
- 'g': Toggle gridFigure Controls:
- Use toolbar for saving, zooming, etc.
- Close window to continue
"""ax_info.text(0.05, 0.4, instructions, transform=ax_info.transAxes, fontsize=10,verticalalignment='top', horizontalalignment='left',bbox=dict(boxstyle='round', facecolor='lightblue', alpha=0.8))plt.tight_layout()# Enable interactive modeplt.ion()plt.show()return fig, axdef plot_unit_cell(self, ax, lattice_vectors):"""Plot unit cell with enhanced visualization"""a1, a2, a3 = lattice_vectorsvertices = np.array([[0, 0, 0],a1,a2,a3,a1 + a2,a1 + a3,a2 + a3,a1 + a2 + a3])edges = [[0, 1], [0, 2], [0, 3],[1, 4], [1, 5],[2, 4], [2, 6],[3, 5], [3, 6],[4, 7], [5, 7], [6, 7]]# Plot edges with different colors for each lattice vectorcolors = ['red', 'green', 'blue', 'red', 'red', 'green', 'green', 'blue', 'blue', 'purple', 'purple', 'purple']for i, edge in enumerate(edges):points = vertices[edge]ax.plot(points[:, 0], points[:, 1], points[:, 2], color=colors[i], linewidth=2.5, alpha=0.7)# Add labels for lattice vectorsax.text(a1[0]/2, a1[1]/2, a1[2]/2, 'a', fontsize=12, color='red', weight='bold')ax.text(a2[0]/2, a2[1]/2, a2[2]/2, 'b', fontsize=12, color='green', weight='bold')ax.text(a3[0]/2, a3[1]/2, a3[2]/2, 'c', fontsize=12, color='blue', weight='bold')def print_lattice_parameters(results):"""Print lattice parameters"""print("\n" + "="*120)print("Lattice Parameters Summary:")print("="*120)print(f"{'Filename':<40} {'a(Å)':<8} {'b(Å)':<8} {'c(Å)':<8} {'alpha':<8} {'beta':<8} {'gamma':<8} {'Volume(ų)':<12}")print("-"*120)for result in results:filename = os.path.basename(result['file'])print(f"{filename:<40} {result['a']:<8.3f} {result['b']:<8.3f} {result['c']:<8.3f} "f"{result['alpha']:<8.3f} {result['beta']:<8.3f} {result['gamma']:<8.3f} {result['volume']:<12.3f}")def main():"""Main function with improved window management"""print("Crystal Strain Tool - Interactive Version")print("=" * 50)# Initializeapp = InteractiveCrystalStrain()# Show current parametersapp.show_current_parameters()# Main loopwhile True:# Get detailed user inputuser_input = app.get_user_input()if not user_input:break# Generate strain seriestry:print(f"\nGenerating strained structures...")results = app.generate_strain_series(user_input)# Show resultsprint(f"Generated {len(results)} strained structures:")print_lattice_parameters(results)# Ask for visualizationroot = tk.Tk()root.withdraw()visualize = messagebox.askyesno("Visualization", "Show 3D structure visualization?")root.destroy()if visualize:# Show original structureprint("Showing original structure... Close the window to continue.")app.plot_structure_3d(title="Original Structure")# Wait for the user to close the windowplt.ioff()plt.show()# Show a strained structureif results:example_idx = len(results) // 2example_result = results[example_idx]strained_lattice = example_result['lattice_vectors']print(f"Showing strained structure: {os.path.basename(example_result['file'])}")print("Close the window to continue.")app.plot_structure_3d(lattice_vectors=strained_lattice,title=f"Strained Structure\n{os.path.basename(example_result['file'])}")plt.ioff()plt.show()# Ask to continueroot = tk.Tk()root.withdraw()continue_choice = messagebox.askyesno("Continue", "Apply another strain?")root.destroy()if not continue_choice:breakexcept Exception as e:root = tk.Tk()root.withdraw()messagebox.showerror("Error", f"Error generating strained structures: {str(e)}")root.destroy()import tracebacktraceback.print_exc()root = tk.Tk()root.withdraw()messagebox.showinfo("Finished", "Program execution completed")root.destroy()if __name__ == "__main__":main()