作者,Evil Genius
没想到公司调整裁员来得这么快这么猛烈,IT组整组被干掉了,财务优化了2个,人人自危了。
还是那句话,做好自己,尽人事听天命吧。
大家还是要好好学习,如此卷的社会常态下,大家不可避免会被卷入这个趋势。
单细胞空间外显子目前还是很需要学习的,但是一切都在基础之上,学会了基础的操作部分,我们会教学写代码的时候如何进行构思,考虑方法的取舍,尽量贴合自己的课题目标,以及如何优化脚本等等。基础真的很重要,是我们书写单细胞空间外显子脚本的基石,现在也成了进入行业和职场的必备技能,大家有条件还是尽量参加一下培训,2025番外--linux、R、python培训 https://mp.weixin.qq.com/s/8E1vYJMNhe5m0DXieBHfzA.
https://mp.weixin.qq.com/s/8E1vYJMNhe5m0DXieBHfzA.
当初我入行生物信息走的是曲线救国的路线,先进入做产品,然后跟着生信大佬学习,一段时间后转到生信,当然,期间也要通过考核,不通过连产品都不能做了,只能走人,现在这种策略很难实施了,而且风险很大,大家如果做生信,还是一步到位最好。
今天我们继续更新脚本,关于高精度的通讯分析问题,大家都知道,CosMx、Xenium等数据每个细胞的转录本数量偏少,基因中位数200很多时候都是奢望,这个时候套用单细胞那样的通讯分析,其实是不合适的。
另一方面,空间通讯需要考虑邻域
我们今天更新这部分
library(Seurat)
library(tidyverse)
library(dplyr)
library(ggplot2)
library(tibble)
### load data objects ###
Lymphoma_data <- readRDS("Lymphoma_data.rds") #This is saved from Figure 1b.
spatial_niche <- readRDS("spatial_niche.rds")
spatial_niche <- column_to_rownames(spatial_niche, var="Barcode")
Lymphoma_data <- AddMetaData(Lymphoma_data, spatial_niche)####li
cell_cell_communication_res = readRDS('cell_cell_communication_res.rds')
####cell-cell communication
Lymphoma.meta <- Lymphoma_data@meta.data
meta_for_cell1 <- select(Lymphoma.meta, c("cell_state","CN_cluster","sample_ID"))
colnames(meta_for_cell1)[1:2] <- c("cell_state1","CN_cluster1")
meta_for_cell1 <- rownames_to_column(meta_for_cell1, var="cell1")
cell_cell_communication_res <- left_join(cell_cell_communication_res, meta_for_cell1, by="cell1")meta_for_cell2 <- select(Lymphoma.meta, c("cell_state","CN_cluster"))
colnames(meta_for_cell2)[1:2] <- c("cell_state2","CN_cluster2")
meta_for_cell2 <- rownames_to_column(meta_for_cell2, var="cell2")
cell_cell_communication_res <- left_join(cell_cell_communication_res, meta_for_cell2, by="cell2")cell_cell_communication_res$cell_pair <- paste0(cell_cell_communication_res$cell1,":",cell_cell_communication_res$cell2)aggretrate <- function(a,b,mol_a,mol_b){data_1 <- filter(cell_cell_communication_res, cell_cell_communication_res$cell_state1==a & cell_cell_communication_res$cell_state2 == b) data_1 <- data_1[,c(paste0(mol_a,".",mol_b,"_a2b"),"cell_pair","cell1","cell2","cell_state1","CN_cluster1","cell_state2","CN_cluster2")]data_2 <- filter(cell_cell_communication_res, cell_cell_communication_res$cell_state1==b & cell_cell_communication_res$cell_state2 == a)data_2 <- data_2[,c(paste0(mol_a,".",mol_b,"_b2a"),"cell_pair","cell1","cell2","cell_state1","CN_cluster1","cell_state2","CN_cluster2")]colnames(data_1)[1] <- "intensity"colnames(data_2)[1] <- "intensity"data <- rbind(data_1, data_2)data_filter <- filter(data, data$CN_cluster1==data$CN_cluster2)data_filter$LR <- paste0(mol_a,"-",mol_b)data_filter$count_ind <- 0data_filter$count_ind[data_filter$intensity > 0] <- 1group_mean <- data_filter %>% group_by(CN_cluster1) %>% summarise(mean_value = mean(intensity))group_mean <- as.numeric(group_mean$mean_value)group_count <- data_filter %>% group_by(CN_cluster1) %>% summarise(count_value = sum(count_ind))group_count_sum <- table(data_filter$CN_cluster1)group_prop <- as.numeric(group_count$count_value/group_count_sum)data_filter$group_mean <- NAdata_filter$group_prop <- NAgroup <- unique(data_filter$CN_cluster1)[order(unique(data_filter$CN_cluster1))]for (i in 1:length(group)) {data_filter$group_mean[data_filter$CN_cluster1== group[i]] <- group_mean[i]data_filter$group_prop[data_filter$CN_cluster1== group[i]] <- group_prop[i]*100}return(data_filter)
}cell_states <- c("C0_Tumor-B","C1_PC_IgG","C2_PC_IgA","C3_Resting-B","C4_PC_IgM","C5_T","C6_TAM_APOE_C1Q","C7_TAM_SPP1","C8_Mac_DUSP1","C9_Mac_CXCL8","C10_Mac_MT2A","C11_FRC","C12_HEV","C13_Endothelial_VWF","C14_VSMC","C15_Stromal_CLU","C16_Stressed","C17_Epithelial","C18_RBC")
agg_table <- function(b, mol_b, mol_a){table <- data.frame()for(i in mol_a){for(j in cell_states){df <- aggretrate(j,b,i,mol_b)df_unique <- unique(select(df, c("CN_cluster1","LR","group_mean","group_prop"))) df_unique$sender <- jtable <- rbind(table, df_unique)}}return(table)
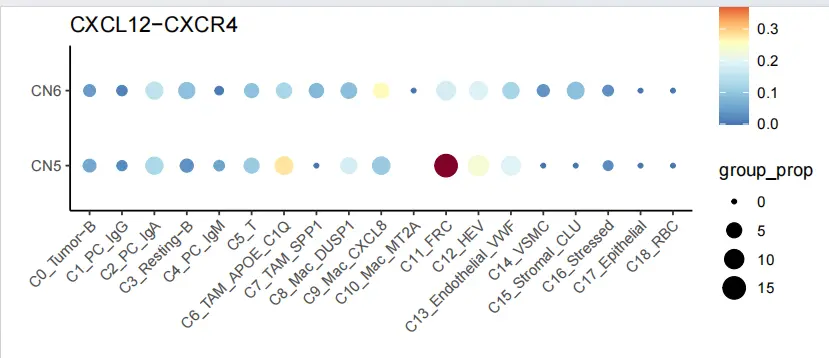
}C0_CXCR4_agg_table <- agg_table("C0_Tumor-B", "CXCR4", "CXCL12")
C0_CXCR4_agg_table$sender <- factor(C0_CXCR4_agg_table$sender, levels = c("C0_Tumor-B","C1_PC_IgG","C2_PC_IgA","C3_Resting-B","C4_PC_IgM","C5_T","C6_TAM_APOE_C1Q","C7_TAM_SPP1","C8_Mac_DUSP1","C9_Mac_CXCL8","C10_Mac_MT2A","C11_FRC","C12_HEV","C13_Endothelial_VWF","C14_VSMC","C15_Stromal_CLU","C16_Stressed","C17_Epithelial","C18_RBC"))ggplot(filter(C0_CXCR4_agg_table, C0_CXCR4_agg_table$CN_cluster1 %in% c("CN5","CN6")), aes(x = sender, y = CN_cluster1, color = group_mean, size = group_prop)) + scale_color_gradientn(values = seq(0,1,0.1),colours = c("#4575b4","#74add1","#abd9e9","#e0f3f8","#ffffbf","#fdae61","#d73027","#800026"))+geom_point(stat = 'identity') + xlab("") + ylab("") + ggtitle("CXCL12-CXCR4") + theme_classic()+theme(axis.text.x = element_text(angle = 45, hjust = 1))
生活很好,有你更好
 https://mp.weixin.qq.com/s/8E1vYJMNhe5m0DXieBHfzA.
https://mp.weixin.qq.com/s/8E1vYJMNhe5m0DXieBHfzA.
