QCResUNet:体素级与受试者级联合的分割质量预测|文献速递-文献分享
Title
题目
QCResUNet: Joint subject-level and voxel-level segmentation qualityprediction
QCResUNet:体素级与受试者级联合的分割质量预测
01
文献速递介绍
医学影像分割质量控制(QC)方法综述与QCResUNet的提出 在现代医学中,医学影像分割通过精准勾勒解剖结构与病变区域,在疾病的准确诊断、监测、治疗方案制定及人群研究中发挥着不可或缺的作用(Garcia-Garcia 等,2017;Litjens 等,2017)。尤其是利用磁共振成像(MRI)将健康与异常解剖结构精准分割为多个类别,是这一过程的关键环节。近年来,基于深度学习的方法在自动分割任务中取得了最先进的性能,包括脑肿瘤(Ronneberger 等,2015;Kamnitsas 等,2017;Isensee 等,2018a,b;Baid 等,2021)和心脏(Tran,2016;Khened 等,2019;Zhou 等,2023)MRI分割。 然而,深度神经网络对数据分布敏感。由于采集协议、对比度、图像质量等因素的差异,将其应用于分布外MRI扫描时,性能易下降。因此,在分割结果用于临床或大规模研究前,需通过质量控制(QC)全面评估。QC工具需实现:逐病例检测严重分割失败、在体素级别定位需修正的分割区域、为下游分析提供质量指标。现有QC方法主要分为四类:基于不确定性估计、基于生成模型、基于反向分类准确率(RCA)、基于回归。 ## 一、现有四类分割QC方法的局限性 ### 1. 基于不确定性估计的方法 这类方法的核心假设是“高不确定性对应低质量分割”(Ng 等,2018;Albà 等,2018;Sander 等,2020 等)。多数研究通过构建不确定性度量,将体素级不确定性聚合为分割质量指标(如Dice相似系数,DSC)的替代指标,但多数替代指标与DSC的相关性较弱(Ng 等,2018;Roy 等,2018;Jungo 等,2020)。此外,体素级不确定性估计存在不可忽略的误差,导致受试者级别的不确定性聚合结果不可靠(Jungo 等,2020)。部分分割方法虽内置QC功能(Kalkhof 和 Mukhopadhyay,2023),但仅适用于基于深度学习的模型,无法评估其他方法生成的分割结果。 ### 2. 基于生成模型的方法 其核心假设是“图像强度与组织标签存在关联”。Grady 等(2012)提出结合手工特征(几何特征、强度特征、梯度特征、比率特征)的高斯混合模型,用于表征分割质量并检测分割失败;Wang 等(2020a)提出变分自编码器(VAE)学习图像与真值分割对的 latent 表示,推理时冻结编码器、微调解码器生成替代分割,再计算查询分割与替代分割的受试者级DSC,但解码器需为每个查询图像单独微调,计算成本高且耗时;Li 等(2022)利用图像到图像翻译技术,提出生成对抗网络(GAN),基于待评估的查询分割掩码生成参考图像,再通过辅助网络(差异分析器)输入原始图像与参考图像,预测图像级和像素级质量。但这类方法仅在心脏MRI分割QC中验证,适用于单一模态和规则形状分割,难以迁移到多模态、肿瘤内组织异质性高的脑肿瘤分割QC场景。 ### 3. 基于反向分类准确率(RCA)的方法 基于RCA框架(Valindria 等,2017),最初用于全身多器官MRI分割QC,后应用于心脏MRI数据集(Robinson 等,2019)。流程包括:(1)选择含真值分割的参考数据集;(2)利用查询图像-分割对训练分割器;(3)用训练后的分割器分割参考数据集图像;(4)将分割器在参考数据集中的最大DSC作为估算结果。虽在全身多器官和心脏MRI分割QC中有效,但依赖代表性参考数据集和图像配准,在脑肿瘤分割中面临挑战——脑肿瘤变异性大,参考数据集难以具备代表性;且与形状、外观稳定的全身多器官/心脏分割不同,脑肿瘤异质性高,难以建立不同受试者肿瘤区域的对应关系。此外,RCA框架仅能预测受试者级DSC,无法在体素级别定位分割误差。 4. 基于回归的方法 这类方法直接预测受试者级DSC。早期研究结合支持向量机回归与手工特征,检测心脏MRI分割失败(Kohlberger 等,2012;Albà 等,2018);Robinson 等(2018)提出卷积神经网络(CNN)回归器,从大型心脏MRI分割数据集中自动提取特征以预测DSC;Kofler 等(2022)提出整体评分,模拟神经放射科专家对分割质量的分级,但专家手动标注整体评分耗时且存在评分者间差异,难以应用于大规模数据集。部分多维回归QC方法(如Fournel 等,2021)通过估算单个2D切片的分割质量指标并整合得到3D预测,但该方法专为DSC预测设计,且无法在体素级别定位分割误差。 ## 二、现有QC方法的共性局限 1. 任务单一性:多数研究仅针对单一分割任务(如全身多器官、心脏MRI分割QC),且未在外部数据集上评估分布外泛化能力。 2. 脑肿瘤针对性不足:针对脑肿瘤MRI分割QC的研究有限,脑肿瘤位置、大小、形状异质性高,增加了QC难度。 3. 质量指标不全面:多数方法仅关注受试者级DSC,未结合轮廓类指标(如归一化表面Dice,NSD),而二者对全面评估分割质量均至关重要(Maier-Hein 等,2024)。 4. 体素级误差定位缺失:可靠的体素级、组织特异性分割误差定位对审计和放射科医生优先修正病例至关重要,但现有研究多忽略此点;仅Li 等(2022)的方法具备相关能力,但仅在有限数据集的心脏分割QC中验证,且仅能生成二值分割误差掩码,无法识别不同组织类别的体素级误差,临床适用性受限。 ## 三、QCResUNet的提出与核心改进 为解决上述局限,本文提出新型深度学习模型QCResUNet,可同时预测受试者级分割质量指标,并在体素级别定位不同组织类别的分割误差。该研究基于前期初步工作(Qiu 等,2023),主要改进包括: 1. 扩展质量评估维度:不仅预测受试者级DSC,还新增NSD指标,并生成对应不同组织类别的二值分割误差图谱,实现更全面的质量评估;同时提出基于注意力的分割误差图谱聚合机制,优化不同组织类别的分割误差勾勒效果。 2. 强化泛化能力验证:在内部数据集(BraTS 2021,1251例)上进行三折交叉验证,在图像质量各异、分割方法不同的分布外数据集上评估,验证模型泛化性。 3. 拓展任务适用性:评估模型在心脏MRI分割QC中的性能,验证其在脑肿瘤之外的更广泛QC任务中的应用潜力。 4. 对比先进方法:将模型与基于RCA(Valindria 等,2017)、基于不确定性(Jungo 等,2020)的主流QC方法对比,全面验证性能优势。 5. 增强可解释性:深入分析模型性能优于其他方法的原因,提升结果可信度。 ## 四、QCResUNet的核心贡献 1. 提出多任务学习框架QCResUNet,可同时预测受试者级DSC与NSD,定位体素级分割误差。 2. 设计基于注意力的分割误差图谱聚合机制,优化不同组织类别的体素级分割误差预测效果。 3. 基于脑肿瘤分割QC任务,通过内部(BraTS 2021三折交叉验证)与外部(WUSM、BraTS-SSA数据集)测试验证模型泛化性,结果表明模型对不同脑肿瘤数据集、不同分割方法生成的分布外样本均有良好适配性。 4. 在心脏MRI分割QC任务中验证模型性能,证明其在脑肿瘤之外的QC任务中的应用潜力。 5. 开源代码(https://github.com/sotiraslab/QCResUNet),助力研究可复现性与领域后续研究。 要不要我帮你整理一份现有QC方法局限性与QCResUNet改进措施的对比表格?
Aastract
摘要
Deep learning has made significant strides in automated brain tumor segmentation from magnetic resonanceimaging (MRI) scans in recent years. However, the reliability of these tools is hampered by the presence ofpoor-quality segmentation outliers, particularly in out-of-distribution samples, making their implementationin clinical practice difficult. Therefore, there is a need for quality control (QC) to screen the quality ofthe segmentation results. Although numerous automatic QC methods have been developed for segmentationquality screening, most were designed for cardiac MRI segmentation, which involves a single modality anda single tissue type. Furthermore, most prior works only provided subject-level predictions of segmentationquality and did not identify erroneous parts segmentation that may require refinement. To address theselimitations, we proposed a novel multi-task deep learning architecture, termed QCResUNet, which producessubject-level segmentation-quality measures as well as voxel-level segmentation error maps for each availabletissue class. To validate the effectiveness of the proposed method, we conducted experiments on assessingits performance on evaluating the quality of two distinct segmentation tasks. First, we aimed to assess thequality of brain tumor segmentation results. For this task, we performed experiments on one internal (BrainTumor Segmentation (BraTS) Challenge 2021, 𝑛 = 1, 251) and two external datasets (BraTS Challenge 2023in Sub-Saharan Africa Patient Population (BraTS-SSA), 𝑛 = 40; Washington University School of Medicine(WUSM), 𝑛 = 175). Specifically, we first performed a three-fold cross-validation on the internal dataset usingsegmentations generated by different methods at various quality levels, followed by an evaluation on theexternal datasets. Second, we aimed to evaluate the segmentation quality of cardiac Magnetic ResonanceImaging (MRI) data from the Automated Cardiac Diagnosis Challenge (ACDC, 𝑛 = 100). The proposed methodachieved high performance in predicting subject-level segmentation-quality metrics and accurately identifyingsegmentation errors on a voxel basis. This has the potential to be used to guide human-in-the-loop feedbackto improve segmentations in clinical settings.
QCResUNet:用于分割质量预测的多任务深度学习架构 近年来,深度学习在基于磁共振成像(MRI)扫描的脑肿瘤自动分割领域取得了显著进展。然而,低质量的分割异常结果(尤其在分布外样本中)影响了这类工具的可靠性,使其难以在临床实践中推广应用。因此,亟需通过质量控制(QC)来筛选分割结果的质量。 尽管目前已有多种自动QC方法用于分割质量筛选,但多数方法是为心脏MRI分割设计的,且仅适用于单一模态和单一组织类型。此外,多数现有研究仅能提供受试者级别的分割质量预测,无法定位可能需要修正的分割错误区域。 为解决这些局限性,我们提出了一种新型多任务深度学习架构——QCResUNet。该架构可生成受试者级别的分割质量评估指标,并为每种可用组织类别生成体素级别的分割误差图谱。 为验证所提方法的有效性,我们通过实验评估了其在两项不同分割任务中的性能。首先,评估脑肿瘤分割结果的质量:实验采用一个内部数据集(2021年脑肿瘤分割挑战赛(BraTS)数据集,样本量(n=1251))和两个外部数据集(撒哈拉以南非洲患者群体脑肿瘤分割挑战赛(BraTS-SSA)数据集,样本量(n=40);华盛顿大学医学院(WUSM)数据集,样本量(n=175))。具体而言,我们先在内部数据集上对不同方法生成的、不同质量水平的分割结果进行三折交叉验证,再在外部数据集上进行评估。其次,评估心脏MRI数据的分割质量:采用自动心脏诊断挑战赛(ACDC)数据集(样本量(n=100))。 实验结果表明,所提方法在预测受试者级别分割质量指标方面表现优异,且能精准定位体素级别的分割错误。该方法有望用于指导“人机协同反馈”,进而在临床场景中优化分割结果。 要不要我帮你提炼这份内容的核心创新点与实验设计总结?
Method
方法
2.1. Dataset
In this study, we used three datasets for evaluation of the QCperformance on brain tumor segmentation. First, we used pre-operativemultimodal MRI scans with gliomas of all grades (WHO Central Nervous System grades 2–4) grades from the BraTS 2021 challenge trainingdataset (𝑛 = 1251). The BraTS dataset is a heterogeneous datasetconsisting of cases from 23 different sites with various levels of qualityand protocols. The BraTS dataset was used for training, validation,and internal testing. Additionally, we used two datasets that are notincluded in the BraTS 2021 dataset (i.e., the BraTS-SSA dataset andthe WUSM dataset) for external testing, allowing for an unbiasedassessment of the generalizability of the proposed method. The BraTSSSA dataset (𝑛 = 40) (Adewole et al., 2023) is an extension of theoriginal BraTS 2021 dataset with patients from Sub-Saharan Africa,which includes lower-quality MRI scans (e.g., poor image contrast andresolution) as well as unique characteristics of gliomas (i.e., suspectedhigher rates of gliomatosis cerebri). The WUSM dataset (𝑛 = 175)was obtained from the retrospective health records of the WashingtonUniversity School of Medicine (WUSM), with a waiver of consent, inaccordance with the Health Insurance Portability and AccountabilityAct, as approved by the Institutional Review Board (IRB) of WUSM(IRB no. PA18-1113). Each subject in all datasets comprised fourmodalities viz. pre-contrast T1-weighted (T1), T2-weighted (T2), postcontrast T1-contrast (T1c), and Fluid attenuated inversion recovery(FLAIR). In addition, multi-class tumor segmentation masks annotatedby experts were also available. Segmentation masks delineated enhancing tumor (ET), non-enhancing tumor core (NCR), and edema (ED)classes. Following standard BraTS procedures, we combined the binaryET, NCR and ED segmentation masks to delineate the whole tumor(WT), tumor core (TC), and enhancing tumor. The WT mask consistsof all tumor tissue classes (i.e., ET, NCR, and ED), while the TC maskcomprises ET and NCR tissue classes.
Scans from the BraTS training and BraTS-SSA datasets were alreadyregistered to the SRI24 anatomical atlas (Rohlfing et al., 2010), resampled to 1-mm3 isotropic resolution and skull-stripped. For consistency,raw MRI scans from WUSM were pre-processed following the sameprotocol using the Integrative Imaging Informatics for Cancer Research:Workflow Automation for Neuro-oncology (Chakrabarty et al., 2022)framework. Subsequently, we z-scored all the skull-stripped scans in theBraTS datasets and the WUSM dataset on a per-scan basis. Finally, scansfrom the entire dataset were cropped to exclude background regionsand then were zero-padded to a common dimension of 160 × 192 × 160using the nnUNet preprocessing pipeline (Isensee et al., 2018b).In evaluating the QC performance on the cardiac segmentationtask, we used the Automated Cardiac Diagnosis Challenge (ACDC)dataset (Bernard et al., 2018), which consists of 100 subjects (200 volumes). Each volume in the ACDC dataset is associated with a multi-classsegmentation mask delineating the left ventricle (LV), myocardium(Myo), and right ventricle (RV). Lastly, each volume was cropped andzero-padded to a common dimension of 16 × 16 × 160 using thennUNet pipeline (Isensee et al., 2018b).
2.1 数据集 本研究采用三个数据集评估脑肿瘤分割的质量控制(QC)性能。首先,使用2021年脑肿瘤分割挑战赛(BraTS 2021)训练数据集(样本量(n=1251)),该数据集包含所有病理级别的胶质瘤(世界卫生组织中枢神经系统分级2-4级)术前多模态磁共振成像(MRI)扫描数据。BraTS数据集具有异质性,涵盖23个不同研究中心的病例,且图像质量与采集协议存在差异,主要用于模型的训练、验证与内部测试。 此外,为无偏评估所提方法的泛化能力,采用两个未纳入BraTS 2021数据集的外部数据集进行测试,分别为BraTS-SSA数据集(样本量(n=40))(Adewole等,2023)与华盛顿大学医学院(WUSM)数据集(样本量(n=175))。其中,BraTS-SSA数据集是BraTS 2021数据集的扩展,病例来源于撒哈拉以南非洲地区,包含质量较低的MRI扫描数据(如图像对比度差、分辨率低),且胶质瘤具有独特特征(如疑似脑胶质瘤病发病率更高);WUSM数据集来自华盛顿大学医学院的回顾性健康记录,经该机构伦理审查委员会(IRB)批准(伦理编号PA18-1113),符合《健康保险流通与责任法案》(HIPAA)相关规定,无需获得受试者知情同意。 所有数据集的每个受试者均包含四种成像模态,分别为T1加权预增强成像(T1)、T2加权成像(T2)、T1加权增强后成像(T1c)及液体衰减反转恢复成像(FLAIR)。同时,数据集还提供由专家标注的多类别肿瘤分割掩码,涵盖增强肿瘤(ET)、非增强肿瘤核心(NCR)与水肿(ED)三类区域。参照BraTS标准流程,将ET、NCR、ED的二值分割掩码组合,进一步划分出全肿瘤(WT)、肿瘤核心(TC)与增强肿瘤区域:其中,WT掩码包含所有肿瘤组织类别(ET、NCR、ED),TC掩码包含ET与NCR两类组织。 BraTS训练数据集与BraTS-SSA数据集的扫描图像已完成与SRI24解剖图谱(Rohlfing等,2010)的配准,重采样至1mm³各向同性分辨率,并进行了颅骨剥离处理。为保证一致性,WUSM数据集的原始MRI扫描图像采用“癌症研究综合影像信息学:神经肿瘤工作流自动化”框架(Chakrabarty等,2022),遵循相同流程完成预处理。随后,对BraTS系列数据集与WUSM数据集中所有经颅骨剥离的扫描图像,按单扫描图像维度进行z分数标准化。最后,采用nnUNet预处理流程(Isensee等,2018b),对所有数据集的扫描图像进行裁剪以去除背景区域,并零填充至统一尺寸160×192×160。 在心脏分割任务的QC性能评估中,采用自动心脏诊断挑战赛(ACDC)数据集(Bernard等,2018),该数据集包含100个受试者(200个容积图像)。每个容积图像均配有多类别分割掩码,标注出左心室(LV)、心肌(Myo)与右心室(RV)区域。最终,通过nnUNet流程(Isensee等,2018b)对每个容积图像进行裁剪与零填充,统一尺寸为16×16×160。 要不要我帮你整理一份各数据集核心信息(用途、样本量、模态、预处理)的汇总表格?
Conclusion
结论
In this work, we proposed QCResUNet, a novel 3D CNN architecturedesigned for automated QC of multi-class tissue segmentation in MRIscans. To the best of our knowledge, this is the first study to providereliable simultaneous subject-level segmentation quality predictionsand voxel-level identification of segmentation errors for different tissueclasses. The results suggest that the proposed method is a promisingapproach for large-scale automated segmentation QC and for guidingclinicians’ feedback for refining segmentation results.A key feature of the proposed method is the multi-task objective.This enabled the proposed method to focus on regions where errorshave occurred, leading to improved performance. This is supportedby the following observations. First, we observed that the CAM ofthe QCResUNet encoder focused more on the regions where the segmentation error occurred compared to ResNet-34 and ResNet-50 (referto Fig. 7). Second, we observed that the supervision from the segmentation error prediction task can in turn guide the DSC and NSDprediction task to prioritize these error-prone regions. As suggestedby the CAM, ResNet-34 and ResNet-50 achieved more accurate DSCand NSD prediction than the UNet, while the UNet performed betterin segmentation error localization. The possible reason behind this isthat the final average pooling layer in the UNet treats each element inthe last feature map equally while ignoring the actual size of tumors. Incontrast, the average pooling in the embedded feature space in ResNetbased methods operates on the abstracted quality feature maps topreserve information, which resulted in better predictive performance.In contrast, the joint optimization of both subject-level and voxellevel predictions by the proposed QCResUNet allows it to combine theadvantages of both ResNet-based models and UNet. As a consequence,QCResUNet can simultaneously localize segmentation errors and assessthe overall quality of the segmentation.
Importantly, the proposed method exhibited high generalizabilitywhen applied to unseen data, surpassing other state-of-the-art segmentation QC methods. This was particularly true for the RCA and UE-basedmethods in the brain tumor segmentation QC task, which exhibitedpoor performance when assessing the quality of segmentation resultsobtained using segmentation methods different than the ones used togenerate training data. The poor generalizability of the RCA methodwas mainly due to the inherent difficulty of obtaining a representativereference dataset for brain tumor segmentation, which is subject tosignificant variability. Such significant variability may violate the underlying assumption of the RCA method that there is at least one samplein the reference dataset that can be successfully segmented given aquery image-segmentation pair (Robinson et al., 2017; Valindria et al.,2017), which is only valid when dealing with healthy anatomies. Inthe case of the cardiac segmentation QC task, which involves healthyanatomy, RCA methods achieved better performance compared to thebrain tumor case when implemented with the atlas-based segmentation method. Similar to RCA, the UE-based QC did not perform wellon subject-level quality prediction as well as localizing segmentationerror in the brain tumor case. This may be attributed to the fact thatthere is inherent variability in uncertainty maps produced by varioussegmentation methods on datasets with different image quality, tumorcharacteristics, etc. While in the cardiac segmentation QC task, whichinvolves healthy anatomy and less variability, the UE-based showedbetter performance. Additionally, as consistent with findings in Jungoet al. (2020), we found that the UE-based method offers limited segmentation error localization. This limitation further hinders its abilityto generalize effectively. Furthermore, the UE-based method can onlybe used to assess segmentations obtained from deep learning models.Unless the deep learning segmentation method directly outputs anestimate of voxel-wise uncertainty, test time estimation of uncertainty(e.g., using MCDropout (Gal and Ghahramani, 2016)) requires accessto the model architecture and weights, which may not be possible formodels deployed in clinical settings.
QCResUNet的核心价值与性能优势分析 本文提出QCResUNet,这是一种新型3D卷积神经网络(CNN)架构,专为MRI扫描中多类别组织分割的自动质量控制(QC)设计。据我们所知,这是首个可同时实现可靠的受试者级别分割质量预测,以及不同组织类别体素级别分割误差识别的研究。结果表明,该方法在大规模自动分割QC,以及指导临床医生反馈以优化分割结果方面极具应用前景。 ## 一、QCResUNet的核心优势:多任务学习目标 多任务学习目标是该方法的关键特征,能让模型聚焦于误差发生区域,进而提升性能,这一点可通过以下观察得到验证: 1. 类激活图谱(CAM)聚焦误差区域:对比ResNet-34和ResNet-50,QCResUNet编码器的CAM能更精准地聚焦于分割误差发生的区域(参见图7)。 2. 误差预测任务反向指导质量预测:分割误差预测任务的监督信号,可反过来引导DSC和NSD预测任务优先关注高误差风险区域。 3. 融合ResNet与UNet的优势:CAM结果显示,ResNet-34和ResNet-50在DSC、NSD预测上比UNet更准确,但UNet在分割误差定位上表现更优。 - UNet的局限在于,其最后的平均池化层对最后一层特征图的每个元素同等对待,忽略了肿瘤的实际大小。 - ResNet类方法则在嵌入特征空间中进行平均池化,操作对象是抽象后的质量特征图,能保留关键信息,因此预测性能更优。 而QCResUNet通过联合优化受试者级别与体素级别预测,融合了ResNet类模型与UNet的优势,可同时实现分割误差定位与分割整体质量评估。 ## 二、QCResUNet的泛化性优势:超越现有先进方法 该方法在未见过的数据上表现出高泛化性,性能超过其他现有先进分割QC方法,尤其在脑肿瘤分割QC任务中,相比基于RCA和UE的方法优势显著——这两类方法在评估“非训练数据所用分割方法”生成的分割结果时,性能较差。 ### 1. 基于反向分类准确率(RCA)方法的泛化性局限 RCA方法泛化性差的主要原因,在于难以获取具有代表性的脑肿瘤分割参考数据集——脑肿瘤存在显著的变异性,这种变异性可能打破RCA方法的核心假设。 RCA方法的假设是:“参考数据集中至少存在一个样本,可通过给定的查询图像-分割对成功分割”(Robinson等,2017;Valindria等,2017),但该假设仅在处理健康解剖结构时成立。 而在心脏分割QC任务中(涉及健康解剖结构),当结合基于图谱的分割方法时,RCA方法的性能会优于脑肿瘤分割场景。 ### 2. 基于不确定性估计(UE)方法的泛化性局限 与RCA类似,基于UE的QC方法在脑肿瘤分割场景中,无论是受试者级别质量预测,还是体素级别分割误差定位,表现均不佳,原因如下: - 不确定性图谱变异性大:不同分割方法在“图像质量、肿瘤特征各异的数据集”上生成的不确定性图谱,存在固有的变异性。 - 健康解剖结构场景下性能较好:仅在心脏分割QC任务(涉及健康解剖结构,变异性较低)中,基于UE的方法才表现出更优性能。 - 误差定位能力有限:与Jungo等(2020)的研究结果一致,我们发现基于UE的方法在分割误差定位上能力有限,这一局限进一步削弱了其有效泛化的能力。 - 临床应用受限:该方法仅能用于评估深度学习模型生成的分割结果。若深度学习分割方法未直接输出体素级不确定性估计,那么测试时的不确定性估计(如使用蒙特卡洛 dropout(Gal和Ghahramani,2016))需要获取模型架构与权重——而在临床部署的模型中,这一点往往难以实现。 要不要我帮你整理一份QCResUNet与RCA、UE方法的核心差异对比表?
Results
结果
4.1. Evaluation of subject-level QC performance
4.1.1. Brain tumor MRI segmentation QC task
The proposed model performed well in the subject-level DSC andNSD prediction across all three brain tumor datasets (Table 4). Specifically, on the BraTS internal testing set with segmentations generatedby nnUNet and nnFormer, the proposed method achieved a small meanMAE of 0.056 and 0.064 for NSD and DSC prediction, respectively.The predicted NSD and DSC also showed a strong correlation withthe corresponding ground truth, achieving Pearson r values of 0.958and 0.968, respectively. Importantly, the proposed method generalizedwell for segmentation produced by different methods (i.e., DeepMedic),which had not been used during training, achieving an average MAE of0.074 and 0.062 for NSD and DSC prediction. Similarly, the predictedNSD and DSC showed a strong correlation with their ground truth withPearson r values of 0.937 and 0.962, respectively.Critically, our method also generalized well to completely unseenexternal BraTS-SSA and WUSM datasets. On the BraTS-SSA dataset,the proposed method achieved an MAE of 0.057 and 0.060 for NSDand DSC prediction, respectively. In addition, the Pearson 𝑟 betweenthe predicted segmentation quality measures and their ground-truthvalues demonstrated a high correlation (NSD 𝑟 = 0.954; DSC 𝑟 = 0.964).Despite containing MRI scans with varying image quality and tumorcharacteristics, the proposed method still generalized well to the BraTSSSA dataset. However, there was a slight drop in performance on theWUSM dataset with a Pearson r of 0.920 and 0.912 for NSD and DSCpredictions, respectively. The MAE for the NSD and DSC prediction onthe WUSM dataset was 0.075 and 0.087, respectively. We conjecturedthis might be attributed to domain shift due to differences in dataacquisition, preprocessing, and the variability of shape and structuresin brain tumors.Importantly, the proposed method outperformed all baseline methods in NSD and DSC prediction tasks. Compared to the three regressionbased QC methods (i.e., UNet, ResNet-34, ResNet-50), QCResUNetimproved the second-best method by 0.8% for NSD prediction and1.5% for DSC prediction in terms of Pearson r values on the BraTSinternal testing set. On the external BraTS-SSA and WUSM datasets,QCResUNet outperformed the second-best method by an average of1.3% and 1.9% in terms of Pearson r, respectively. A paired t-testconfirmed that this improvement was statistically significant comparedto all three regression-based methods baseline (Table 4). In addition,the proposed method outperformed multi-dimensional regression-basedmethod (Fournel et al., 2021) in subject-level DSC prediction by anaverage of 56.3%, 58.8%, and 180% on internal testing, external BraTSSSA, and external WUSM datasets, respectively (Table 4). We hypothesize that the inferior performance of multi-dimensional regressionbased methods, compared to other regression-based approaches, isattributed to the aggregation of 2D DSCs across slices without accounting for tumor sizes. As a consequence, each slice contributes equallyto the aggregation of 2D DSCs. However, the variability in tumorsizes within a single 3D subject or across different subjects presents asignificant challenge in accurately aggregating them for subject-levelDSC prediction. The proposed approach exhibited more evenly distributed DSC prediction errors across different quality levels comparedto all the baseline methods (refer to Figs. 3 and 4), demonstrating asmaller standard deviation in MAE (see Table 4). Furthermore, ourQCResUNet offers computational efficiency comparable to ResNet50and slightly surpasses that of UNet, especially in terms of FLOPs (seemore computational benchmarking in Appendix F).The proposed method demonstrated a strong performance gain overthe state-of-the-art RCA and UE-based QC methods (Table 4). The performance of the RCA and the UE-based method after hyper-parametertuning was in line with previous works Robinson et al. (2017) andJungo et al. (2020). In the internal BraTS testing set, the proposedmethod improved the average Pearson r of NSD predictions by 48.5%and DSC predictions by 22.1% compared to RCA and UE-based QC,respectively. A similar trend was observed on the external datasets,with the proposed method improving Pearson 𝑟 by an average of 50.9%and 48.2% compared to RCA and UE-based QC, respectively. Moreover,the proposed method achieved a significant reduction in the averageMAE of predicting NSD and DSC compared to the RCA and UE-basedmethods for all results viz. internal testing results (0.292, 0.126 vs.0.064), BraTS-SSA results (0.281, 0.157 vs. 0.059), and WUSM results(0.275, 0.179 vs. 0.081).
4.1 受试者级别质量控制(QC)性能评估 ## 4.1.1 脑肿瘤磁共振成像(MRI)分割QC任务 所提模型在三个脑肿瘤数据集上的受试者级别Dice相似系数(DSC)与归一化表面Dice(NSD)预测任务中均表现优异(表4)。具体而言,在采用nnUNet和nnFormer生成分割结果的BraTS内部测试集上,该方法对NSD和DSC预测的平均绝对误差(MAE)较低,分别为0.056和0.064;预测的NSD与DSC还与对应真值呈强相关性,皮尔逊相关系数(Pearson r)分别达到0.958和0.968。 重要的是,该方法对训练过程中未使用的分割方法(如DeepMedic)生成的结果也具备良好泛化性:对NSD和DSC预测的平均MAE分别为0.074和0.062,且预测值与真值的相关性依然较强,Pearson r分别为0.937和0.962。 关键在于,该方法对完全未见过的外部数据集(BraTS-SSA和WUSM)同样表现出良好泛化性。在BraTS-SSA数据集上,其对NSD和DSC预测的MAE分别为0.057和0.060,预测的分割质量指标与真值的Pearson r均体现出高相关性(NSD r=0.954;DSC r=0.964)。尽管该数据集包含图像质量与肿瘤特征各异的MRI扫描数据,所提方法仍能稳定适配。 不过,在WUSM数据集上,模型性能出现小幅下降:NSD和DSC预测的Pearson r分别为0.920和0.912,MAE分别为0.075和0.087。我们推测,这可能是由数据采集、预处理流程的差异,以及脑肿瘤形状与结构的变异性导致的领域偏移所致。 值得注意的是,所提方法在NSD和DSC预测任务中均优于所有基准方法。相较于三种基于回归的QC方法(UNet、ResNet-34、ResNet-50),在BraTS内部测试集上,QCResUNet对NSD和DSC预测的Pearson r,分别比第二优方法提升0.8%和1.5%;在外部数据集BraTS-SSA和WUSM上,其Pearson r平均分别比第二优方法提升1.3%和1.9%。配对t检验证实,相较于这三种基于回归的基准方法,该提升具有统计学显著性(表4)。 此外,在受试者级别DSC预测任务中,所提方法还优于基于多维回归的方法(Fournel等,2021):在内部测试集、外部BraTS-SSA数据集、外部WUSM数据集上,性能分别平均提升56.3%、58.8%和180%(表4)。我们认为,基于多维回归的方法性能较差(相较于其他基于回归的方法),原因在于其未考虑肿瘤大小,直接对各切片的2D DSC进行聚合——每个切片对2D DSC聚合的贡献权重相同,而单个3D受试者内部或不同受试者间的肿瘤大小存在差异,这为准确聚合2D DSC以实现受试者级别DSC预测带来了巨大挑战。 与所有基准方法相比,所提方法在不同质量水平下的DSC预测误差分布更均匀(参见图3和图4),MAE的标准差更小(见表4)。此外,QCResUNet的计算效率与ResNet-50相当,且略优于UNet,尤其在浮点运算次数(FLOPs)方面(更多计算基准测试详见附录F)。 相较于当前最先进的基于反向分类准确率(RCA)和不确定性估计(UE)的QC方法,所提方法同样体现出显著的性能优势(表4)。经超参数调优后,基于RCA和UE的方法性能与已有研究(Robinson等,2017;Jungo等,2020)结果一致。在BraTS内部测试集上,所提方法对NSD和DSC预测的平均Pearson r,相较于基于RCA和UE的QC方法分别提升48.5%和22.1%;在外部数据集上也观察到类似趋势,其Pearson r平均分别提升50.9%和48.2%。 此外,在所有结果中(内部测试结果:0.292、0.126 vs 0.064;BraTS-SSA结果:0.281、0.157 vs 0.059;WUSM结果:0.275、0.179 vs 0.081),相较于基于RCA和UE的方法,所提方法对NSD和DSC预测的平均MAE均实现显著降低。 要不要我帮你整理一份QCResUNet与各基准方法在脑肿瘤QC任务上的核心性能指标对比表?
Figure
图
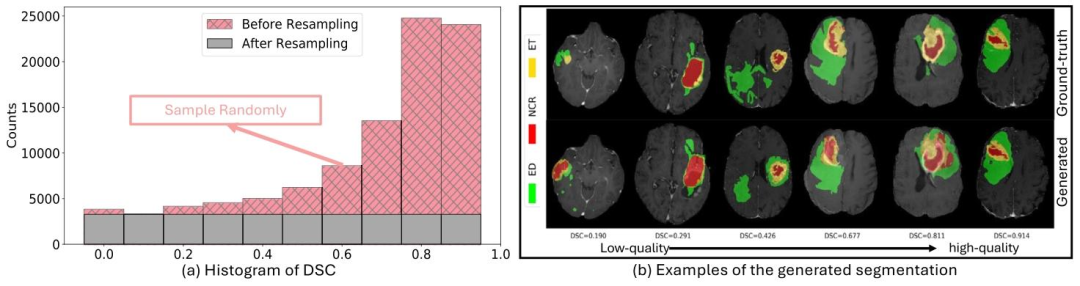
Fig. 1. Data generation for the brain tumor segmentation QC task: (a) The histogram of the DSC distribution in the generated BraTS training dataset before and after applyingthe resampling strategy. (b) Visual examples of the generated segmentation dataset ranging from low-quality to high-quality
图1 脑肿瘤分割质量控制(QC)任务的数据生成 (a)采用重采样策略前后,生成的脑肿瘤分割挑战赛(BraTS)训练数据集中Dice相似系数(DSC)分布的直方图。 (b)生成的分割数据集从低质量到高质量的可视化示例。
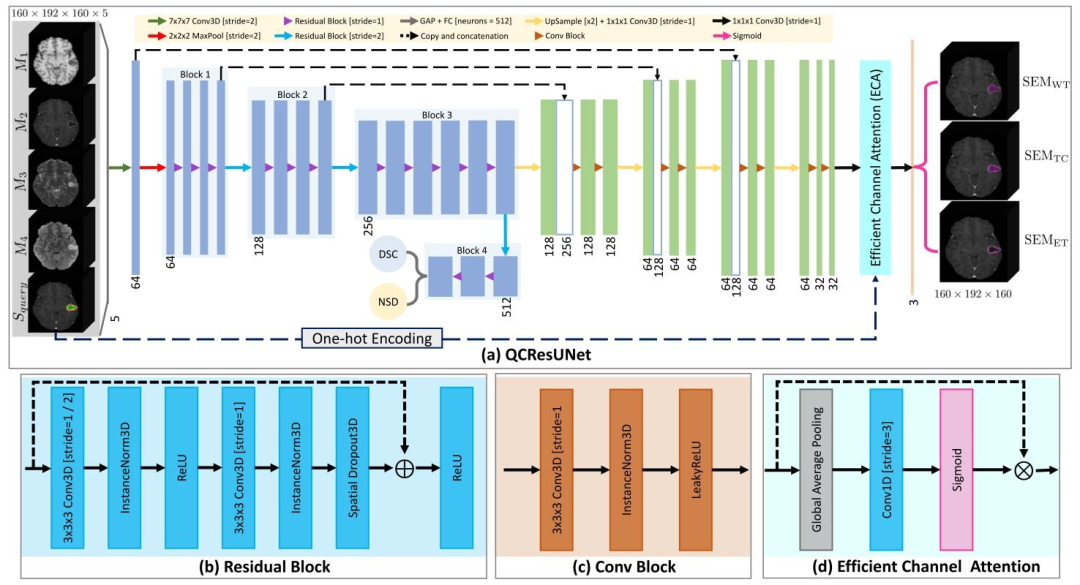
Fig. 2. (a) The proposed QCResUNet model is a U-shaped neural network that takes as input 𝑚 imaging modalities ({𝑀1 ,𝑀2 , … ,𝑀𝑚 }) and a multi-class segmentation mask tobe evaluated (𝑆**𝑞𝑢𝑒𝑟𝑦). It generates three outputs: the subject-level segmentation-quality metrics DSC (DSC𝑝𝑟𝑒𝑑 ) and NSD (NSD𝑝𝑟𝑒𝑑 ) as well as a collection of 𝐶 binary voxel-levelsegmentation error maps {SEMtissue𝑐} 𝐶 𝑐=1, each for each tissue class. In the case of brain tumor segmentation task, which is demonstrated as an example in this figure, QCResUNettakes as input four imaging modalities ({𝑀1 ,𝑀2 ,𝑀3 ,𝑀4 }) and a query segmentation 𝑆𝑞𝑢𝑒𝑟𝑦 that delineates WT, TC, and ET. It produces subject-level DSC and NSD, along with threebinary SEMs corresponding to the segmentation error masks for WT, TC, and ET tissue classes. Note that the depicted image sizes as well as the number and size of convolutionfilters are specific to the brain tumor segmentation tasks. Subpanel figures (b) (c), and (d) depict the Residual Block employed in the encoder of QCResUNet, the ConvolutionalBlock used in its decoder, and the Efficient Channel Attention (ECA) used for multiclass SEM aggregation, respectively. The abbreviations in the figure: Conv3D = 3D convolutionallayer, GAP = global average pooling, FC = fully connected layer, LeakyReLU = leaky rectified linear unit, and One-hot Encoding = one-hot encodes the multi-class 𝑆𝑞𝑢𝑒𝑟𝑦 to acollection of binary masks.
图2 (a)所提QCResUNet模型为U型神经网络,输入为(m)种成像模态({M₁, M₂, …, Mₘ})与待评估的多类别分割掩码(S₍query₎)。该模型生成三类输出:受试者级分割质量指标Dice相似系数(DSC₍pred₎)、归一化表面Dice(NSD₍pred₎),以及(C)个二值体素级分割误差图谱集合{SEM₍tissue₎^c}((c=1)至(C)),每个图谱对应一个组织类别。本图以脑肿瘤分割任务为例进行展示:QCResUNet的输入为四种成像模态({M₁, M₂, M₃, M₄})与标注全肿瘤(WT)、肿瘤核心(TC)、增强肿瘤(ET)的查询分割掩码(S₍query₎);输出为受试者级DSC与NSD,以及三个分别对应WT、TC、ET组织类别的二值分割误差图谱(SEM)。注:图中所示图像尺寸及卷积滤波器的数量与大小,均为脑肿瘤分割任务专用参数。 子图(b)、(c)、(d)分别展示了QCResUNet编码器中使用的残差块(Residual Block)、解码器中使用的卷积块(Convolutional Block),以及用于多类别分割误差图谱(SEM)聚合的高效通道注意力(ECA)模块。 图中缩写说明:Conv3D=3D卷积层,GAP=全局平均池化,FC=全连接层,LeakyReLU=泄漏整流线性单元,One-hot Encoding=独热编码(将多类别查询分割掩码S₍query₎编码为二值掩码集合)。
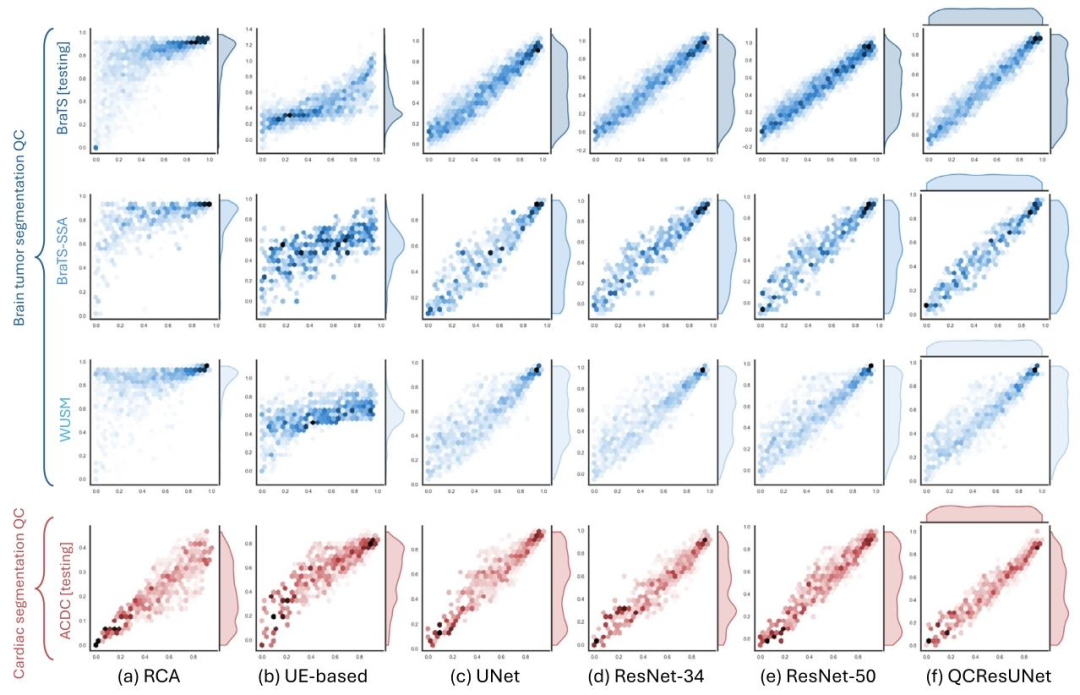
Fig. 3. Scatter plots of the ground-truth (x-axis) and the predicted DSC (y-axis) for the proposed method and all the baseline methods in the internal BraTS testing, externalBraTS-SSA, WUSM datasets, and ACDC internal testing (rows). Results for (a) RCA; (b) UE-based; (c) UNet; (d) ResNet-34; (e)ResNet-50; and (f) QCResUNet are reported in differentcolumns. The proposed method generalized well to external datasets and consistently showed superior performance compared to all baseline methods. The UE-based method showedthe worst performance compared to all other methods.
图3 所提方法与所有基准方法在各数据集上的真值DSC(x轴)与预测DSC(y轴)散点图 行对应不同数据集:内部BraTS测试集、外部BraTS-SSA数据集、WUSM数据集、ACDC内部测试集;列对应不同方法的结果:(a)基于反向分类准确率(RCA)的方法;(b)基于不确定性估计(UE)的方法;(c)UNet;(d)ResNet-34;(e)ResNet-50;(f)QCResUNet。 结果显示,所提方法对外部数据集具有良好的泛化性,且相较于所有基准方法始终表现出更优性能;其中,基于不确定性估计(UE)的方法相较于其他所有方法表现最差。
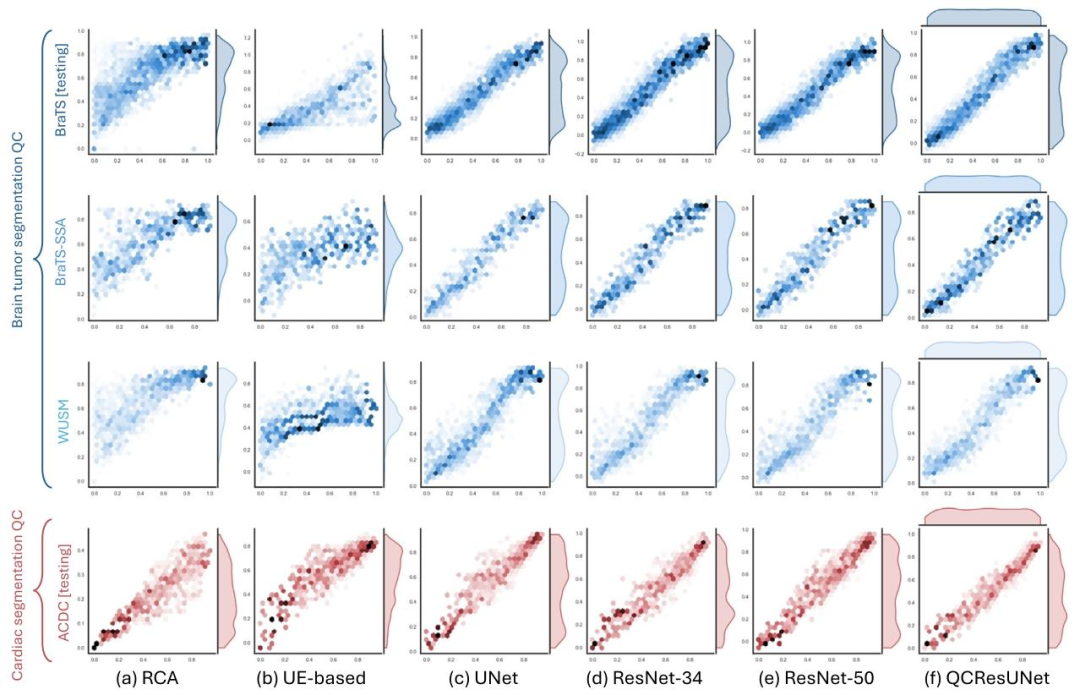
Fig. 4. Scatter plots of the ground-truth (x-axis) and the predicted NSD (y-axis) for the proposed method and all the baseline methods in the internal BraTS testing, externalBraTS-SSA, WUSM datasets, and ACDC internal testing (rows). Results for (a) RCA; (b) UE-based; (c) UNet; (d) ResNet-34; (e)ResNet-50; and (f) QCResUNet are reported in differentcolumns. The proposed method generalized well to external datasets and consistently showed superior performance compared to all baseline methods. The UE-based method showedthe worst performance compared to all other methods.
图4 所提方法与所有基准方法在各数据集上的真值NSD(x轴)与预测NSD(y轴)散点图 行对应不同数据集:内部BraTS测试集、外部BraTS-SSA数据集、WUSM数据集、ACDC内部测试集;列对应不同方法的结果:(a)基于反向分类准确率(RCA)的方法;(b)基于不确定性估计(UE)的方法;(c)UNet;(d)ResNet-34;(e)ResNet-50;(f)QCResUNet。 结果显示,所提方法对外部数据集具有良好的泛化性,且相较于所有基准方法始终表现出更优性能;其中,基于不确定性估计(UE)的方法相较于其他所有方法表现最差。
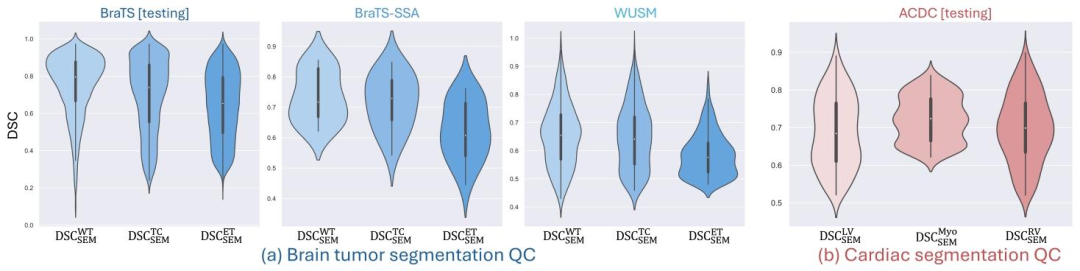
Fig. 5. The distribution of the DSC (DSCSEM) between the predicted segmentation error map and the corresponding ground truth for (a) brain tumor segmentation QC task and (b)cardiac segmentation QC task. The proposed QCResUNet accurately localized segmentation errors in terms of DSCSEM for all tissue classes across all datasets (refer toSection 4.2for detailed discussion), demonstrating good generalization
图5 预测分割误差图谱与对应真值之间的Dice相似系数(DSC₍SEM₎)分布 (a)脑肿瘤分割质量控制(QC)任务;(b)心脏分割QC任务。 结果显示,所提QCResUNet模型在所有数据集的所有组织类别上,均能通过DSC₍SEM₎指标精准定位分割误差(详见4.2节讨论),体现出良好的泛化性。
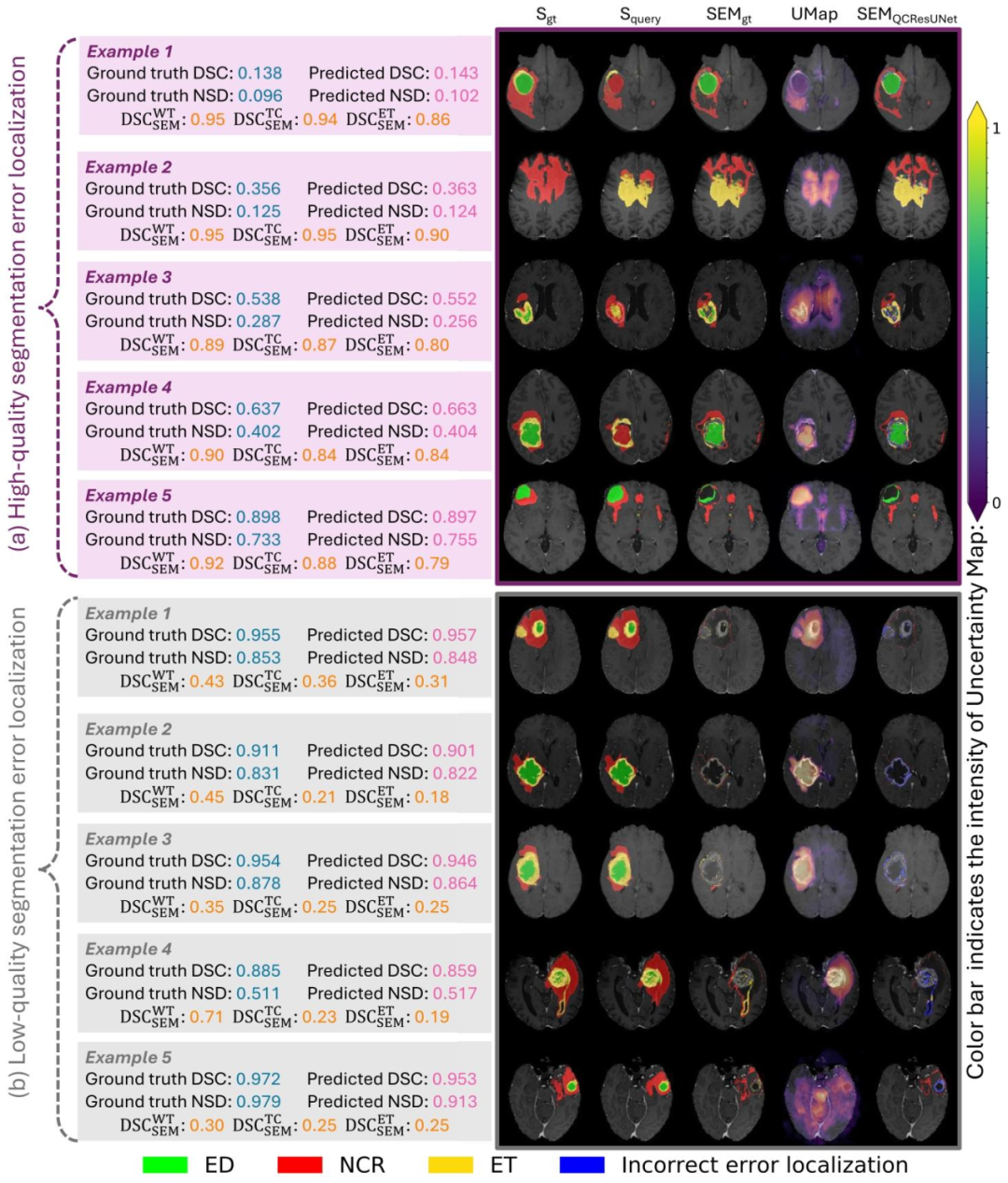
Fig. 6. Examples showcasing the performance of the proposed method versus baseline methods on the brain tumor segmentation QC task: (a) high-quality segmentation errorlocalization and (b) low-quality segmentation error localization. The color bar in the figure indicates the intensity of the uncertainty map (UMap). We observed that the proposedmethod showed better segmentation error localization than the uncertainty map. The error localization was better when dealing with low-quality query segmentation in contrastto higher-quality ones. This may be attributed to the fact that detecting few errors at the boundaries of high-quality segmentations is challenging. We kindly direct the readersto Appendix D for details on how the visualization in this figure was obtained.
图6 脑肿瘤分割质量控制(QC)任务中所提方法与基准方法的性能展示示例 (a)高质量分割的误差定位;(b)低质量分割的误差定位。 图中颜色条代表不确定性图谱(UMap)的强度。结果显示,所提方法的分割误差定位效果优于不确定性图谱;且相较于高质量查询分割,该方法在处理低质量查询分割时,误差定位效果更优。这可能是因为在高质量分割中,检测边界处的少量误差存在较大挑战性。关于本图可视化结果的获取方式,详见附录D。
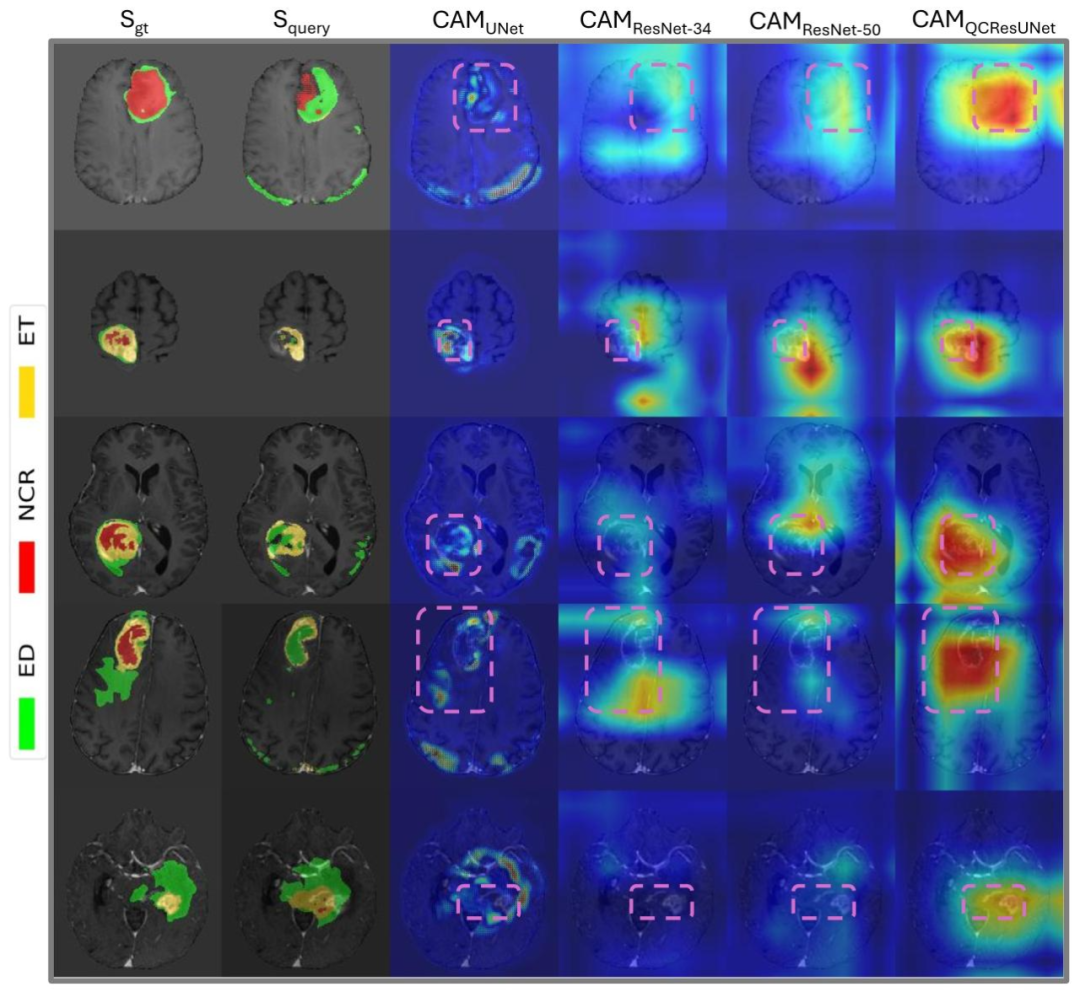
Fig. 7. Examples of the class activation maps produced by Grad-CAM for different methods, with regions of interest (ROIs) highlighted in purple boxes. The CAMs of UNet,ResNet-34, and ResNet-50 were generated based on the last convolutional feature maps. The CAMs of the proposed QCResUNet were generated from the last convolutional featuremap of the ResNet encoder. We observed that the proposed QCResUNet demonstrated improved localization performance compared to the other baselines in terms of the CAM.This may be attributed to the multi-task learning framework of the proposed method (see detailed discussion in Section 4.4)
图7 不同方法通过梯度加权类激活映射(Grad-CAM)生成的类激活图谱(CAM)示例 图中紫色方框标注了感兴趣区域(ROI)。其中,UNet、ResNet-34和ResNet-50的类激活图谱(CAM)基于各自最后一层卷积特征图生成;所提QCResUNet的类激活图谱(CAM)则基于其ResNet编码器的最后一层卷积特征图生成。 我们观察到,从类激活图谱(CAM)来看,所提QCResUNet的定位性能优于其他基准方法。这可能得益于所提方法的多任务学习框架(详见4.4节的详细讨论)。
Table
表
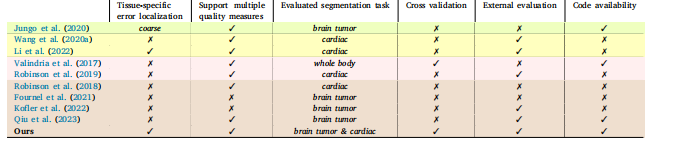
Table 1A detailed comparison of recent segmentation QC methods chosen from four different categories, including UE-based , generative-model-based , RCA-based , andregression-based methods. We chose recent representative methods from each category.
表1 近期分割质量控制(QC)方法的详细对比 该对比涵盖四类方法,包括基于不确定性估计(UE-based)、基于生成模型(generative-model-based)、基于回归系数分析(RCA-based)和基于回归(regression-based)的方法。我们从每类方法中选取了近期具有代表性的方法进行分析。

Table 2The parameters of image transformations used in SegGen
表2 SegGen中所用图像变换的参数
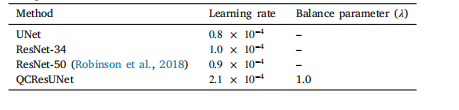
Table 3The optimal hyperparameter settings for different models (i.e., UNet, ResNet-34, ResNet-50, QCResUNet) obtained from performing a random search usingRaytune. Please refer to Appendix C for the details of Raytune hyperparametertuning
表3 不同模型(即UNet、ResNet-34、ResNet-50、QCResUNet)的最优超参数设置 该最优超参数设置通过使用Raytune工具进行随机搜索获得。有关Raytune超参数调优的详细信息,请参见附录C。
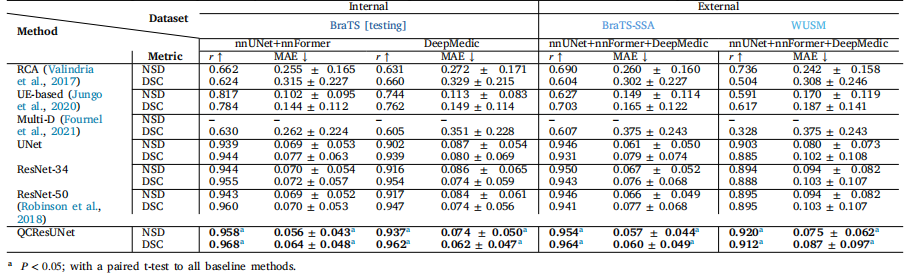
Table 4The subject-level QC performance on the brain tumor MRI segmentation task was evaluated on the internal BraTS testing dataset, as well as the independent BraTS-SSA, andWUSMs datasets with segmentations generated by nnUNet, nnFormer, and DeepMedic. The best metrics within each column are highlighted in bold
表4 脑肿瘤磁共振成像(MRI)分割任务的受试者级别质量控制(QC)性能评估结果 评估基于内部脑肿瘤分割挑战赛(BraTS)测试数据集,以及独立的撒哈拉以南非洲脑肿瘤分割挑战赛(BraTS-SSA)数据集和华盛顿大学医学院(WUSM)数据集;所用分割结果均由nnUNet、nnFormer和DeepMedic三种方法生成。每列中的最优指标以粗体标注。
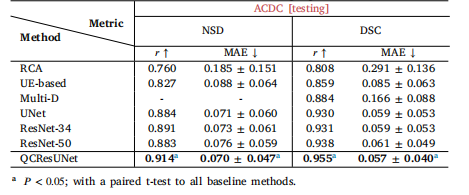
Table 5The subject-level QC performance on the cardiac MRI segmentation task was evaluatedon the internal ACDC testing set with segmentations produced by nnUNer andnnFormer. The best metrics within each column are highlighted in bold.
表5 心脏磁共振成像(MRI)分割任务的受试者级别质量控制(QC)性能评估结果 评估基于内部自动心脏诊断挑战赛(ACDC)测试集;所用分割结果由nnUNet和nnFormer两种方法生成。每列中的最优指标以粗体标注。
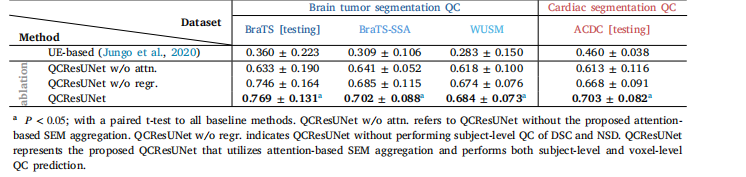
Table 6Comparison of voxel-level segmentation QC Performance of the proposed method to baseline methods in terms of DSCSEM. DSCSEM forthe brain tumor MRI segmentation QC task is computed as the average of DSCWT SEM, DSCTC SEM, and DSCET SEM. While DSCSEM for cardiacMRI segmentation QC task is computed as the average of DSCLV SEM, DSCMyo SEM, and DSCRV SEM. The best metrics within each column arehighlighted in bold.
表6 所提方法与基准方法在体素级别分割质量控制(QC)性能上的对比(以分割误差图谱Dice相似系数(DSC₍SEM₎)为评价指标) 其中,脑肿瘤磁共振成像(MRI)分割QC任务的DSC₍SEM₎计算方式为:全肿瘤分割误差图谱Dice相似系数(DSC₍WT SEM₎)、肿瘤核心分割误差图谱Dice相似系数(DSC₍TC SEM₎)与增强肿瘤分割误差图谱Dice相似系数(DSC₍ET SEM₎)的平均值;心脏MRI分割QC任务的DSC₍SEM₎计算方式为:左心室分割误差图谱Dice相似系数(DSC₍LV SEM₎)、心肌分割误差图谱Dice相似系数(DSC₍Myo SEM₎)与右心室分割误差图谱Dice相似系数(DSC₍RV SEM₎)的平均值。每列中的最优指标以粗体标注。
