R包fastWGCNA - 快速执行WGCNA分析和下游分析可视化
- 最新版本: 1.0.0
- 可以对着视频教程学习和使用:然而还没录呢, 关注B站等我更新

R包介绍
开发背景
- WGCNA是转录组或芯片表达谱数据常用得分析, 可用来鉴定跟分组或表型相关得模块基因和核心基因
- 但其步骤非常之多, 每次运行起来很是费劲, 但需要修改的参数并不多
- 所以完全可以编写成一站式流程, 直接从表达谱和表型得到所有结果
- 另外得到模块基因后, 后续还有PPI和Cytoscape的分析
功能简介
- 一站式实现WGCNA, 只需要输入表达谱和分组或表型文件, 即可完成分析
- 保存各种结果文件, 如每个模块基因、hub基因、显著性结果、MM/GS评分等等
- 可视化内容: 散点图、相关性热图重绘、相关性蝴蝶图
- 下游分析: Cytoscape的MCODE和cytoHubba结果的提取和可视化
安装
-
没有安装包的需要付费获取安装包:
- 直接添加微信转账100, 发送fastR(依赖包)和fastWGCNA的压缩包
- 如果购买过R包fastGEO, 可以半价购买fastWGCNA包!
- 很多资料后面都会慢慢涨价, 早买会便宜很多!
- 购买后可进入交流群,后续版本更新和问题答疑都在群里进行
-
R包fastGEO目前售价调整为100, 以前为20, 逐步涨价到50, 现在是100
-
两包合购限时优惠150, 限时多久看心情
-
所有R包随着功能增加价格必然会上调, 早买早享受, 且免费获取最新版本
# 依赖包
install_if_missing <- function(pkg, repos = getOption("repos")) {if (!requireNamespace(pkg, quietly = TRUE)) {install.packages(pkg, repos = repos)}
}install_if_missing("WGCNA")
install_if_missing("linkET")
install_if_missing("dplyr")
install_if_missing("pdftools")
install_if_missing("UpSetR")
install_if_missing("VennDiagram")
install_if_missing("gridExtra")
# 如果不存在fastR包或版本过低则进行安装
file_pkgs = grep("^fastR.*\\.gz$", dir(), value = TRUE)
if(!require("fastR", quietly = TRUE) || packageVersion("fastR") < strsplit(file_pkgs, "_|\\.tar")[[1]][2]){install.packages(file_pkgs, repos = NULL, upgrade = FALSE, force = TRUE)
}
# 如果不存在fastWGCNA包或版本过低则进行安装
file_pkgs = grep("^fastWGCNA.*\\.gz$", dir(), value = TRUE)
if(!require("fastWGCNA", quietly = TRUE) || packageVersion("fastWGCNA") < strsplit(file_pkgs, "_|\\.tar")[[1]][2]){install.packages(file_pkgs, repos = NULL, upgrade = FALSE, force = TRUE)
}
========================================欢迎使用 fastWGCNA (版本 1.0.0)- 作者: 生信摆渡- 微信: bioinfobaidu1- 帮助文档: help(package = fastWGCNA)
========================================
功能演示
library(fastWGCNA)
# 准备数据, 没有购买fastGEO的可以联系我购买或跳过此步
# 此步仅是数据准备, 也可以自行准备, 然后运行 run_WGCNA
library(fastGEO)
========================================欢迎使用 fastGEO (版本 1.7.0)- 作者: 生信摆渡- 微信: bioinfobaidu1- 帮助文档: help(package = fastGEO)
========================================
run_WGCNA
-
一站式运行WGCNA, 需要输入表达谱(expM)和分组向量(group)或表型数据框(pheno)
-
其他重要参数
- dir: 输出文件夹
- case_name: 实验组的名称
- pheno_name: 表型的名称, 会在图中显示
- ntop: 选取方差最大的前多少基因构建网络, 越少运行速度越快。当模块数或结果基因较少时, 可以调多一点, 默认5000
- npower: 软阈值计算的最大值, 默认是20, 一般就够了, 样本太少时可能需要调到30才会满足构建网络的需求
- cut: 去除离群样本的聚类数的高度, 大于该高度的会被去除, 默认不去
- soft: 选择的软阈值, 默认使用推荐的软阈值, 也可手动指定
- zlim: 相关性热图的相关性范围, 当相关性不高时, 范围调窄点会更好看些
- minModuleSize: 每个模块最小的基因数, 默认30, 最低了
- deepSplit: 数值越大(通常0-4),模块划分越精细,会产生更多更小的模块, 默认2
- reassignThreshold: 基因重新分配的阈值, 影响模块的纯度,较高的阈值可能保留更多模糊归属的基因, 默认0
- mergeCutHeight: 基于模块间的相关性进行合并,值越大,合并的模块越多, 默认0.25
- corr_cutoff: 判定模块显著性的相关系数, 默认为0, 只要P值<0.05就行, 最低阈值
- pvalue_cutoff: 判定模块显著性的P值, 默认0.05
- MM_cutoff: MM阈值, 默认0.8
- GS_cutoff: GS阈值, 默认0.3
-
如果没有得到显著的模块, 可以调整以下参数:
- ntop调大, 最大为nrow(expM), 即所有基因
- deepSplit调大
- mergeCutHeight调小
-
如果没有得到hub基因或太少, 可以调整以下参数:
- MM调低,最小为0.3
- GS调低,最小为0.3
obj = download_GEO("GSE30999", out_dir = "test/00_GEO_data_GSE30999")
INFO [2025-08-27 11:17:17] Step1: Download GEO data ...
INFO [2025-08-27 11:17:17] Querying dataset: GSE30999 ...- Use local curl- Found 1 GPL- Found 170 samples, 39 metas.- Writting sample_anno to test/00_GEO_data_GSE30999/GSE30999_sample_anno.csv - Normalize between arrays ...- Successed, file save to test/00_GEO_data_GSE30999/GSE30999_GPL570.RData.
INFO [2025-08-27 11:17:44] Step2: Annotate probe GPL570 ...
INFO [2025-08-27 11:17:45] Use built-in annotation file ...
- All porbes matched!
- All porbes annotated!
INFO [2025-08-27 11:17:46] Removing duplicated genes by method: max ...
INFO [2025-08-27 11:18:29] Done.
gruop_list = get_GEO_group(obj, group_name = "source_name_ch1", "NL" = "Control", "LS" = "Psoriasis")
expM = gruop_list$expM
group = gruop_list$group
hd(expM)
Type: data.frame Dim: 22881 × 170 GSM767976 GSM767978 GSM767980 GSM767982 GSM767984
A1BG 2.085353 2.079059 2.086824 2.081765 2.082029
A1BG-AS1 3.634412 2.953882 4.084853 3.958588 4.157088
A1CF 2.433706 2.395676 2.432235 2.381176 2.414382
A2M 13.489853 12.870176 12.957765 13.594529 13.878676
A2M-AS1 5.659882 4.673235 4.218735 5.400971 6.311412
table(group)
groupControl Psoriasis 85 85
# expM和group可自行准备
WGCNA_res = run_WGCNA(expM, group, case_name = "Psoriasis", pheno_name = "Psoriasis", dir = "test/GSE30999_WGCNA")
Select top 5000 genes with largest variation ...Check data integrity ...This a good data, nothing to do!Detect outlier samples ...Show_outlier samplesRemove 0 sample(s)Cluster with phenoDetermine soft threshold ...Warning message:
"executing %dopar% sequentially: no parallel backend registered"Not specify soft threshold, using estimated: 6Scale independenceMean connectivityBuild modules ...Plot Cluster dendrogram ...Plot eigengene dendrogram ...Show correlation and P-value between modules and phenos ...Plot butterfly corrplot ...Convert pdf to png ...
- 输出对象
str(WGCNA_res)
List of 12$ datExpr : num [1:170, 1:5000] 2.27 8.36 2.68 2.62 9.24 .....- attr(*, "dimnames")=List of 2.. ..$ : chr [1:170] "GSM767976" "GSM767978" "GSM767980" "GSM767982" ..... ..$ : chr [1:5000] "SERPINB4" "S100A12" "TCN1" "XIST" ...$ datTraits :'data.frame': 170 obs. of 2 variables:..$ Psoriasis: num [1:170] 0 0 0 0 0 0 0 0 0 0 .....$ Control : num [1:170] 1 1 1 1 1 1 1 1 1 1 ...$ MEs :'data.frame': 170 obs. of 14 variables:..$ MEblue : num [1:170] -0.0477 -0.169 -0.1571 -0.1265 -0.0532 .....$ MEgreen : num [1:170] 0.0133 0.0231 -0.0146 -0.1468 -0.2191 .....$ MEturquoise : num [1:170] -0.0726 -0.0644 -0.0859 -0.1298 -0.0848 .....$ MEred : num [1:170] 0.0146 -0.0218 0.1047 0.0307 -0.0513 .....$ MEgreenyellow: num [1:170] 0.08038 -0.00806 -0.00954 -0.03715 -0.00169 .....$ MEyellow : num [1:170] 0.0887 0.1067 0.0628 0.0196 -0.0385 .....$ MEbrown : num [1:170] -0.00425 -0.06434 -0.064 0.04502 0.04749 .....$ MEpink : num [1:170] -0.0199 -0.0351 0.0287 0.1011 0.1063 .....$ MEsalmon : num [1:170] 0.0711 -0.0173 0.0537 0.02 0.0896 .....$ MEtan : num [1:170] 0.1622 0.0444 0.0503 0.1119 0.1766 .....$ MEmagenta : num [1:170] 0.0472 0.037 0.0825 -0.0614 -0.0482 .....$ MEblack : num [1:170] -0.07746 0.04469 -0.01769 -0.00823 -0.03659 .....$ MEpurple : num [1:170] 0.0446 0.0721 -0.0119 -0.0994 -0.1468 .....$ MEgrey : num [1:170] 0.1167 0.0228 0.076 0.1295 0.0814 ...$ all_module_genes:List of 14..$ blue : chr [1:1082] "ABCB1" "ABCC10" "ABHD14A-ACY1" "ABHD2" .....$ green : chr [1:156] "AIF1" "ALOX5AP" "AMICA1" "B3GNT7" .....$ turquoise : chr [1:2295] "A2ML1" "A2MP1" "AAMDC" "AASS" .....$ red : chr [1:147] "ABCA13" "ACAA2" "ACADM" "ACAT2" .....$ greenyellow: chr [1:51] "ACAD8" "AF007147" "AFAP1L1" "AMIGO2" .....$ yellow : chr [1:202] "ABCA6" "ABI3BP" "ACKR4" "ACVRL1" .....$ brown : chr [1:288] "ABAT" "ACCS" "ACTG1P4" "ADCY10P1" .....$ pink : chr [1:113] "AACSP1" "AB488780" "ABO" "ACACB" .....$ salmon : chr [1:37] "ABLIM3" "ACTA1" "ACTC1" "ACTG2" .....$ tan : chr [1:42] "ACVR1C" "ADIPOQ" "AIFM2" "AQP7" .....$ magenta : chr [1:68] "ANGPTL7" "CAPN8" "CBLN2" "COL11A1" .....$ black : chr [1:134] "ADAT1" "AK091729" "ALOX5" "AP006216.10" .....$ purple : chr [1:51] "AADAC" "ACER1" "AF086294" "ASPHD2" .....$ grey : chr [1:334] "ABCC4" "abParts" "ABTB2" "AC007362.3" ...$ signif_summary :List of 2..$ Psoriasis:'data.frame': 14 obs. of 4 variables:.. ..$ corr : num [1:14] 0.353 0.399 0.915 -0.467 -0.343 ..... ..$ pvalue: num [1:14] 2.40e-06 6.96e-08 2.87e-68 1.35e-10 4.68e-06 ..... ..$ Ngene : int [1:14] 1082 156 2295 147 51 202 288 113 37 42 ..... ..$ Sig : chr [1:14] "POS" "POS" "POS" "NEG" .....$ Control :'data.frame': 14 obs. of 4 variables:.. ..$ corr : num [1:14] -0.353 -0.399 -0.915 0.467 0.343 ..... ..$ pvalue: num [1:14] 2.40e-06 6.96e-08 2.87e-68 1.35e-10 4.68e-06 ..... ..$ Ngene : int [1:14] 1082 156 2295 147 51 202 288 113 37 42 ..... ..$ Sig : chr [1:14] "NEG" "NEG" "NEG" "POS" ...$ moduleLabels : Named num [1:5000] 1 1 1 0 0 6 1 1 0 1 .....- attr(*, "names")= chr [1:5000] "SERPINB4" "S100A12" "TCN1" "XIST" ...$ moduleColors : chr [1:5000] "turquoise" "turquoise" "turquoise" "grey" ...$ geneTree :List of 7..$ merge : int [1:4999, 1:2] -15 -40 -39 -71 -63 -4 -5 -89 -459 -106 .....$ height : num [1:4999] 0.146 0.152 0.153 0.158 0.166 .....$ order : int [1:5000] 4355 9 128 2563 4125 712 4990 1436 1489 2812 .....$ labels : NULL..$ method : chr "average"..$ call : language fastcluster::hclust(d = as.dist(dissTom), method = "average")..$ dist.method: NULL..- attr(*, "class")= chr "hclust"$ all_module_genes:List of 14..$ blue : chr [1:1082] "ABCB1" "ABCC10" "ABHD14A-ACY1" "ABHD2" .....$ green : chr [1:156] "AIF1" "ALOX5AP" "AMICA1" "B3GNT7" .....$ turquoise : chr [1:2295] "A2ML1" "A2MP1" "AAMDC" "AASS" .....$ red : chr [1:147] "ABCA13" "ACAA2" "ACADM" "ACAT2" .....$ greenyellow: chr [1:51] "ACAD8" "AF007147" "AFAP1L1" "AMIGO2" .....$ yellow : chr [1:202] "ABCA6" "ABI3BP" "ACKR4" "ACVRL1" .....$ brown : chr [1:288] "ABAT" "ACCS" "ACTG1P4" "ADCY10P1" .....$ pink : chr [1:113] "AACSP1" "AB488780" "ABO" "ACACB" .....$ salmon : chr [1:37] "ABLIM3" "ACTA1" "ACTC1" "ACTG2" .....$ tan : chr [1:42] "ACVR1C" "ADIPOQ" "AIFM2" "AQP7" .....$ magenta : chr [1:68] "ANGPTL7" "CAPN8" "CBLN2" "COL11A1" .....$ black : chr [1:134] "ADAT1" "AK091729" "ALOX5" "AP006216.10" .....$ purple : chr [1:51] "AADAC" "ACER1" "AF086294" "ASPHD2" .....$ grey : chr [1:334] "ABCC4" "abParts" "ABTB2" "AC007362.3" ...$ signif_summary :List of 2..$ Psoriasis:'data.frame': 14 obs. of 4 variables:.. ..$ corr : num [1:14] 0.353 0.399 0.915 -0.467 -0.343 ..... ..$ pvalue: num [1:14] 2.40e-06 6.96e-08 2.87e-68 1.35e-10 4.68e-06 ..... ..$ Ngene : int [1:14] 1082 156 2295 147 51 202 288 113 37 42 ..... ..$ Sig : chr [1:14] "POS" "POS" "POS" "NEG" .....$ Control :'data.frame': 14 obs. of 4 variables:.. ..$ corr : num [1:14] -0.353 -0.399 -0.915 0.467 0.343 ..... ..$ pvalue: num [1:14] 2.40e-06 6.96e-08 2.87e-68 1.35e-10 4.68e-06 ..... ..$ Ngene : int [1:14] 1082 156 2295 147 51 202 288 113 37 42 ..... ..$ Sig : chr [1:14] "NEG" "NEG" "NEG" "POS" ...$ sinif_genes :List of 2..$ Psoriasis: chr [1:4413] "A2ML1" "A2MP1" "AACSP1" "AAMDC" .....$ Control : chr [1:4413] "A2ML1" "A2MP1" "AACSP1" "AAMDC" ...$ hub_genes :List of 14..$ blue :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ green :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ turquoise :'data.frame': 174 obs. of 2 variables:.. ..$ MM: num [1:174] 0.95 0.969 0.943 0.956 0.957 ..... ..$ GS: num [1:174] 0.923 0.953 0.928 0.912 0.936 .....$ red :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ greenyellow:'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ yellow :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ brown :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ pink :'data.frame': 2 obs. of 2 variables:.. ..$ MM: num [1:2] 0.863 0.865.. ..$ GS: num [1:2] 0.803 0.82..$ salmon :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ tan :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ magenta :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ black :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ purple :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0) ..$ grey :'data.frame': 0 obs. of 2 variables:.. ..$ MM: num(0) .. ..$ GS: num(0)
-
输出目录
-
01_all_gene_each_model: 每个模块的所有基因
-
02_phenotype_summary: 显著性统计表, 可用作后续筛选
-
03_sinificant_genes: 对应相关系数和p值下所有显著模块的基因
-
04_MM_GS_scatter: 每个模块
-
前面常规的图就不多解释了, 全都是发表级别的图
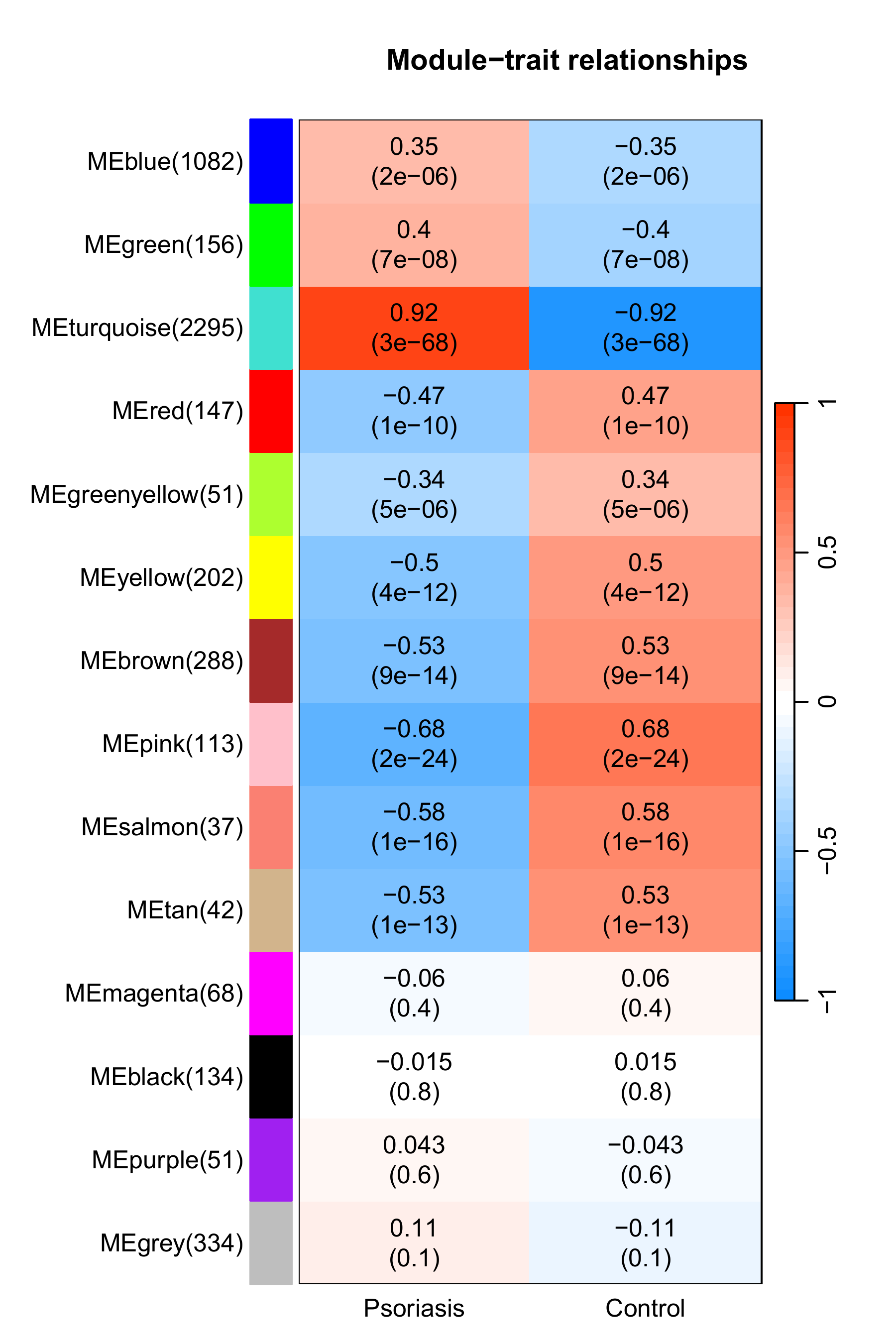
-
相关性热图增加了每个模块的基因数
-
这很重要不是吗? 是我们决定选择哪个模块或判断结果可不可用的关键因素。
-
相关性蝴蝶图
-
相关性的另外一种展示, 还怪好看的
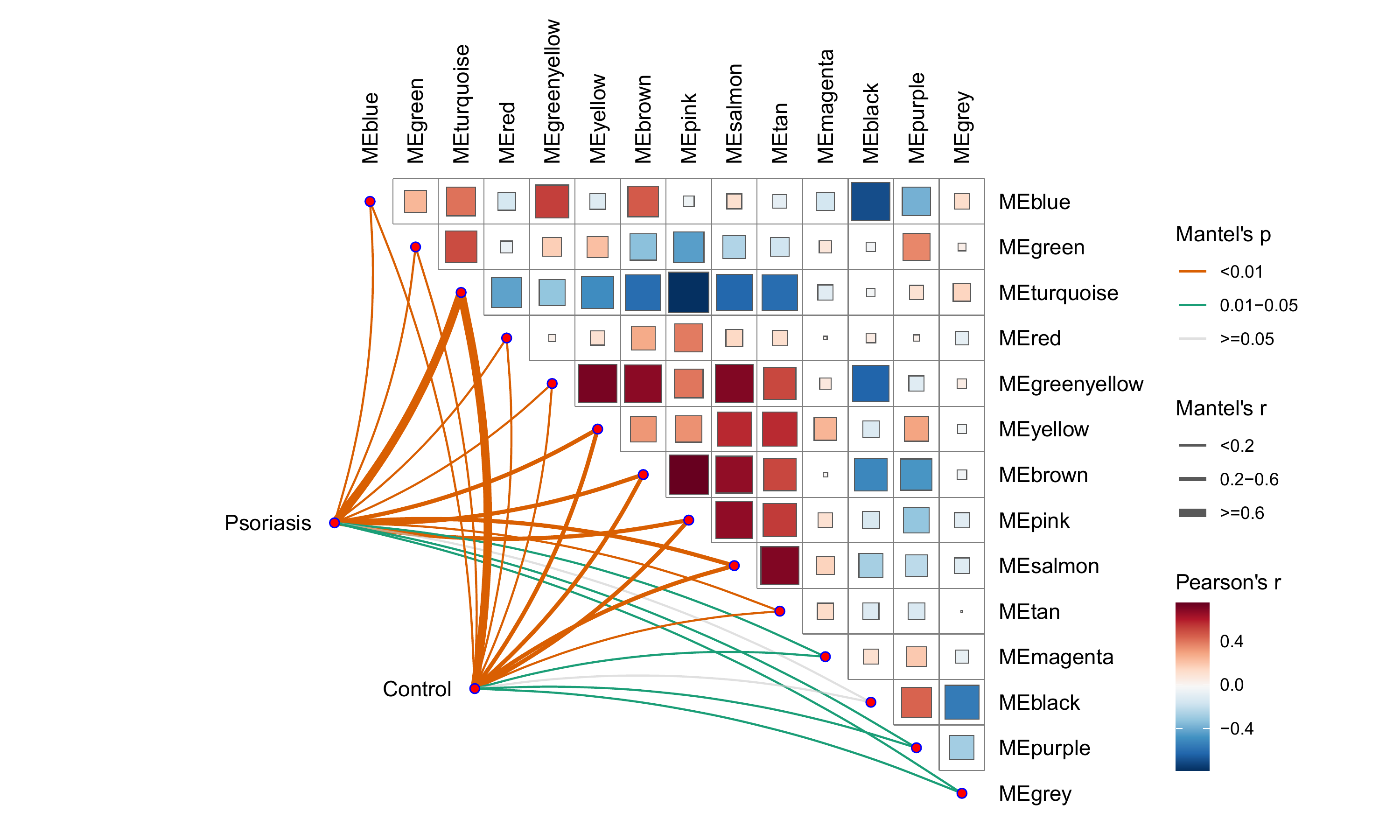
-
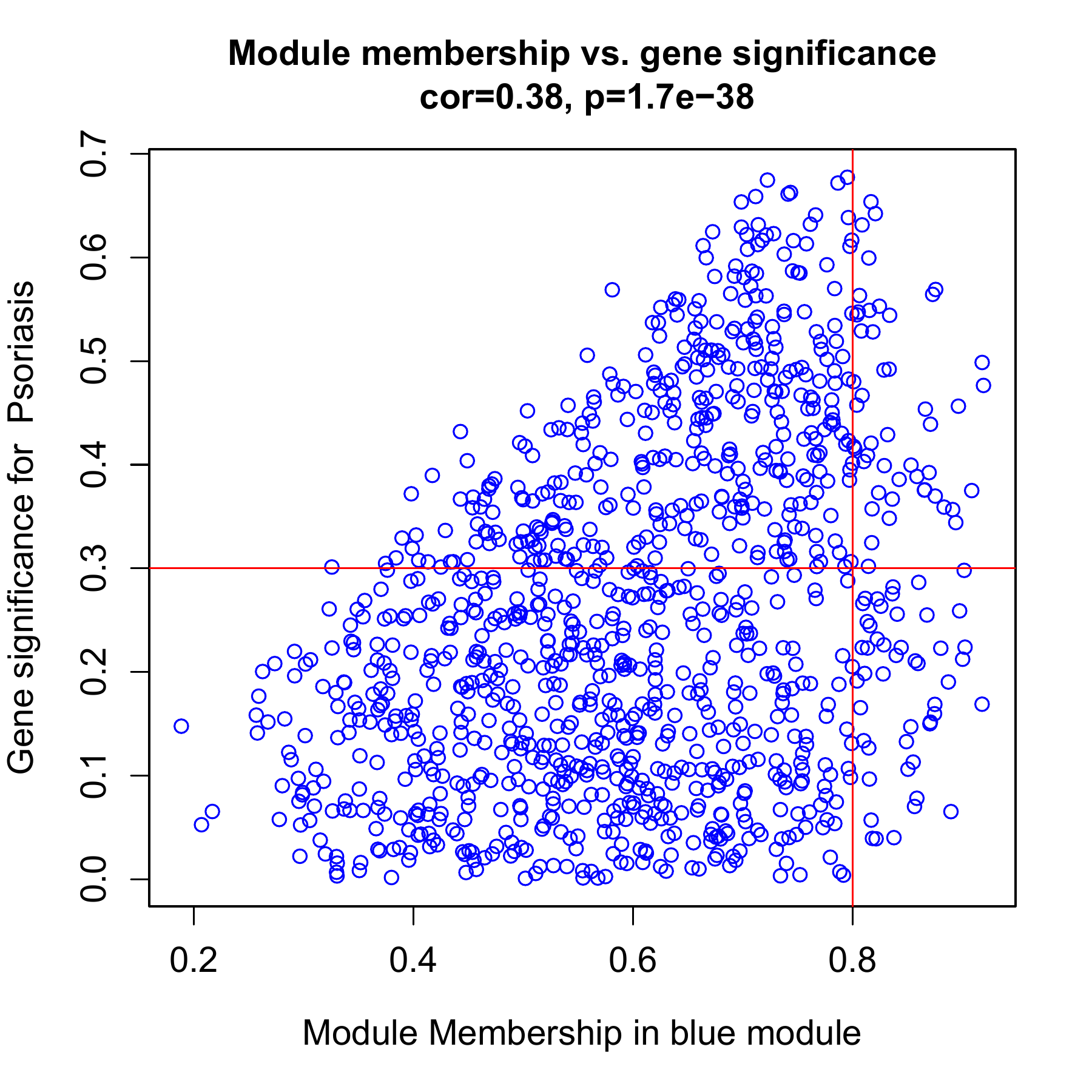
MM MS相关性图
-
如果进行了hub筛选, 这个图是必须要放的, 每个模块一个图
extractMCODE
- 对Cytoscape MCODE插件输出结果进行提取和分析
- 输出原始表格、每一子网络的基因集及其GO富集分析结果
extractMCODE("./test/PPI_MCODE.txt", runGO = FALSE)
$clusterT
A data.frame: 3 × 5
Cluster Score Nodes Edges IDs
<int> <dbl> <int> <int> <chr>
1 5.429 8 19 CDK2, PDGFB, VEGFC, CCND2, NOTCH1, KIT, CCND1, EPHA2
2 3.000 3 3 PKP2, KCNH2, TMEM43
3 3.000 3 3 HBA1, HBB, HBA2
$cluster_list
$Cluster1'CDK2''PDGFB''VEGFC''CCND2''NOTCH1''KIT''CCND1''EPHA2'
$Cluster2'PKP2''KCNH2''TMEM43'
$Cluster3'HBA1''HBB''HBA2'
# 对每一CLuster富集分析, 耗时
mcode_res = extractMCODE("./test/PPI_MCODE.txt")
Loading packages ...Running ONTOLOGY: ALL ...782 terms enrichedLoading packages ...Running ONTOLOGY: ALL ...143 terms enrichedLoading packages ...Running ONTOLOGY: ALL ...55 terms enriched
str(mcode_res$egoL)
List of 3$ Cluster1:'data.frame': 782 obs. of 10 variables:..$ ONTOLOGY : chr [1:782] "BP" "BP" "BP" "BP" .....$ ID : chr [1:782] "GO:1904238" "GO:0033674" "GO:0060326" "GO:0071902" .....$ Description: chr [1:782] "pericyte cell differentiation" "positive regulation of kinase activity" "cell chemotaxis" "positive regulation of protein serine/threonine kinase activity" .....$ GeneRatio : chr [1:782] "3/8" "5/8" "5/8" "4/8" .....$ BgRatio : chr [1:782] "11/21288" "346/21288" "375/21288" "140/21288" .....$ pvalue : num [1:782] 5.74e-09 5.93e-08 8.86e-08 1.23e-07 1.27e-07 .....$ p.adjust : num [1:782] 5.74e-06 2.00e-05 2.00e-05 2.00e-05 2.00e-05 .....$ qvalue : num [1:782] 1.62e-06 5.63e-06 5.63e-06 5.63e-06 5.63e-06 .....$ geneID : chr [1:782] "PDGFB/NOTCH1/EPHA2" "PDGFB/CCND2/KIT/CCND1/EPHA2" "PDGFB/VEGFC/NOTCH1/KIT/EPHA2" "PDGFB/CCND2/KIT/CCND1" .....$ Count : int [1:782] 3 5 5 4 5 5 5 5 4 3 ...$ Cluster2:'data.frame': 143 obs. of 10 variables:..$ ONTOLOGY : chr [1:143] "BP" "BP" "BP" "BP" .....$ ID : chr [1:143] "GO:0086005" "GO:0086091" "GO:0086002" "GO:0086003" .....$ Description: chr [1:143] "ventricular cardiac muscle cell action potential" "regulation of heart rate by cardiac conduction" "cardiac muscle cell action potential involved in contraction" "cardiac muscle cell contraction" .....$ GeneRatio : chr [1:143] "2/3" "2/3" "2/3" "2/3" .....$ BgRatio : chr [1:143] "37/21288" "44/21288" "63/21288" "86/21288" .....$ pvalue : num [1:143] 8.81e-06 1.25e-05 2.58e-05 4.83e-05 4.94e-05 .....$ p.adjust : num [1:143] 0.000669 0.000669 0.000921 0.001057 0.001057 .....$ qvalue : num [1:143] 4.61e-05 4.61e-05 6.34e-05 7.28e-05 7.28e-05 .....$ geneID : chr [1:143] "PKP2/KCNH2" "PKP2/KCNH2" "PKP2/KCNH2" "PKP2/KCNH2" .....$ Count : int [1:143] 2 2 2 2 2 2 2 2 2 2 ...$ Cluster3:'data.frame': 55 obs. of 10 variables:..$ ONTOLOGY : chr [1:55] "BP" "BP" "BP" "BP" .....$ ID : chr [1:55] "GO:0015670" "GO:0015671" "GO:0015669" "GO:0019755" .....$ Description: chr [1:55] "carbon dioxide transport" "oxygen transport" "gas transport" "one-carbon compound transport" .....$ GeneRatio : chr [1:55] "3/3" "3/3" "3/3" "3/3" .....$ BgRatio : chr [1:55] "15/21288" "15/21288" "22/21288" "26/21288" .....$ pvalue : num [1:55] 2.83e-10 2.83e-10 9.58e-10 1.62e-09 2.80e-09 .....$ p.adjust : num [1:55] 5.24e-09 5.24e-09 1.18e-08 1.50e-08 2.07e-08 .....$ qvalue : num [1:55] 2.98e-10 2.98e-10 6.72e-10 8.51e-10 1.18e-09 .....$ geneID : chr [1:55] "HBA1/HBB/HBA2" "HBA1/HBB/HBA2" "HBA1/HBB/HBA2" "HBA1/HBB/HBA2" .....$ Count : int [1:55] 3 3 3 3 3 3 3 3 3 3 ...
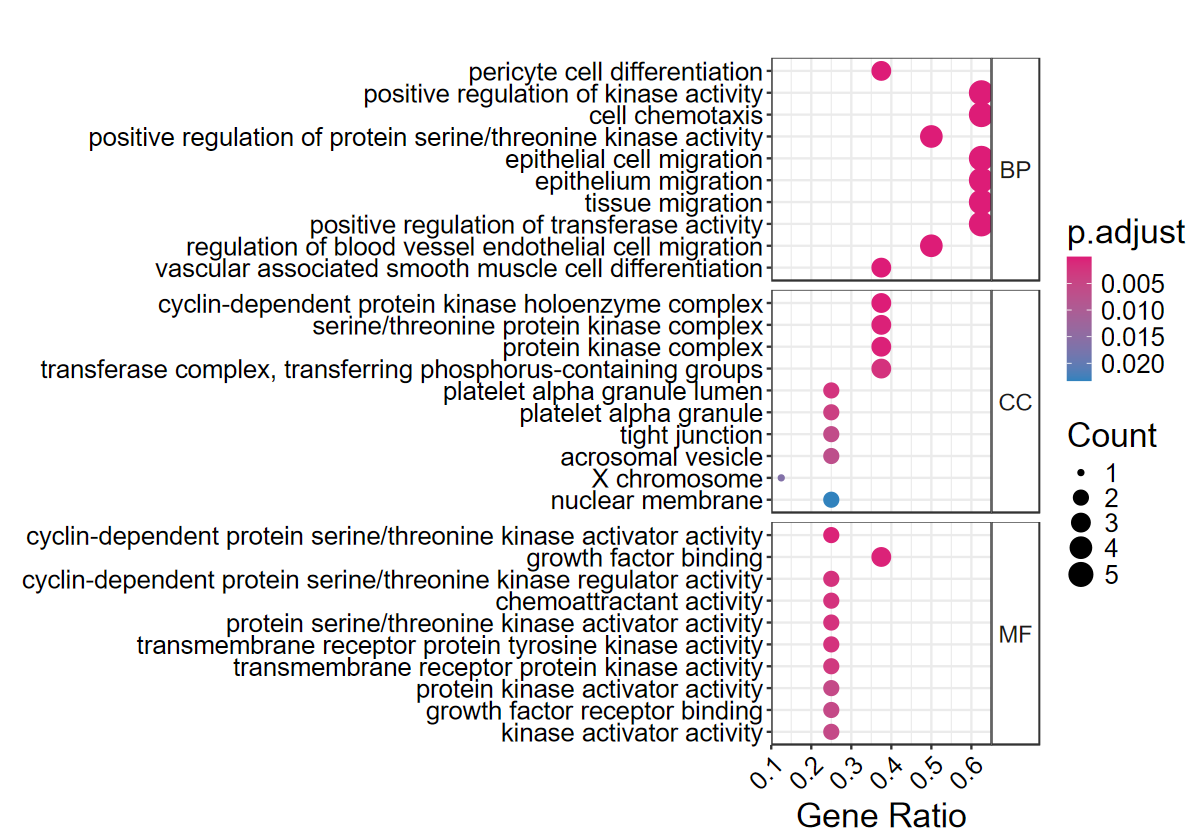
# 简单看下气泡图
set_image(10, 7)
dotplotGO(mcode_res$egoL$Cluster1)
plot_cytohubba_intersection
- 对Cytoscape cytoHubb 插件输出结果进行提取和分析
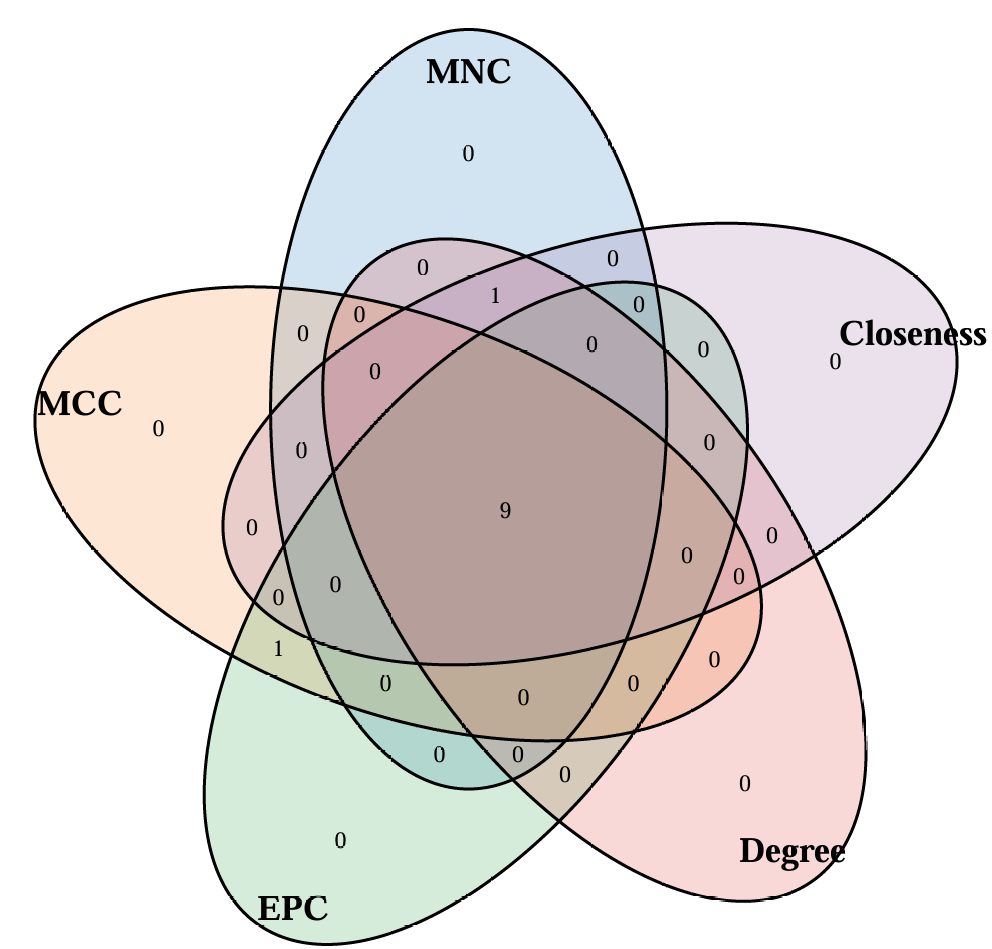
- 提取常用的5种算法的 top N 个基因取交集, 并绘制韦恩图和upset图
cytohubba_res = plot_cytohubba_intersection("./test/PPI_CytoHubba.csv", out_name = "./test/PPI_CytoHubba")
cytohubba_res
$top_genes
$top_genes$MNC[1] "XDH" "IL1B" "HMOX1" "PLAT" "PPARG" "HMOX2" "PRKCB" [8] "BLVRA" "POR" "SULT1A3"$top_genes$MCC[1] "XDH" "IL1B" "HMOX1" "PPARG" "PLAT" "HMOX2" "BLVRA" "PRKCB" "POR"
[10] "PTGES"$top_genes$EPC[1] "XDH" "IL1B" "HMOX1" "PPARG" "PLAT" "HMOX2" "PRKCB" "BLVRA" "POR"
[10] "PTGES"$top_genes$Degree[1] "XDH" "IL1B" "HMOX1" "PLAT" "PPARG" "HMOX2" "PRKCB" [8] "BLVRA" "POR" "SULT1A3"$top_genes$Closeness[1] "XDH" "IL1B" "HMOX1" "PLAT" "PPARG" "HMOX2" "PRKCB" [8] "BLVRA" "POR" "SULT1A3"$common_genes
[1] "BLVRA" "HMOX1" "HMOX2" "IL1B" "PLAT" "POR" "PPARG" "PRKCB" "XDH" $files_generated
[1] "./test/PPI_CytoHubba_upset.pdf"
[2] "./test/PPI_CytoHubba_venn.pdf"
[3] "./test/PPI_CytoHubba_common_genes.csv"
- upset图:
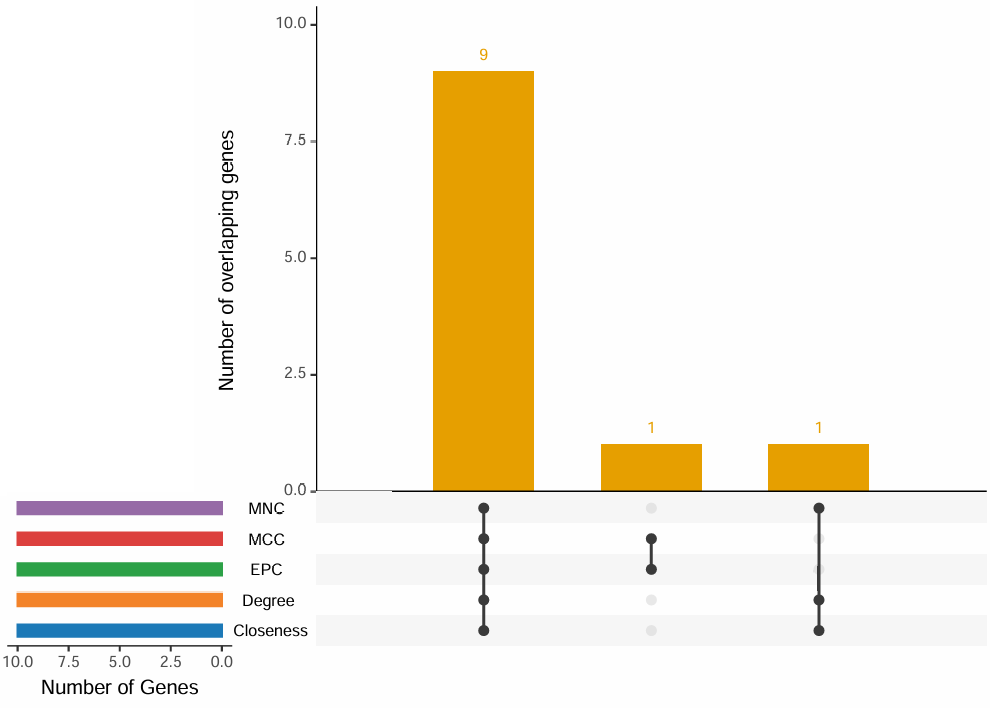
- 韦恩图:
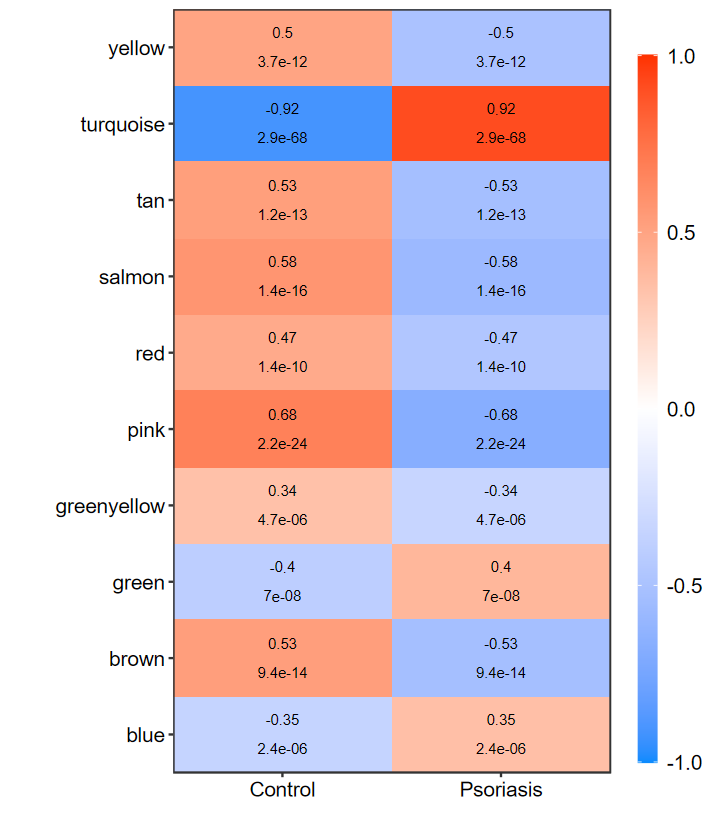
heatmap_replot
- 重新绘制相关性热图, 用于当模块数过多时
- 不大好看, 几年前写的了, 后面再优化
- 已发表的论文用过我做的这个图了, 关键信息都有了
# 只显示显著的
set_image(6, 7)
heatmap_replot(WGCNA_res$signif_summary)
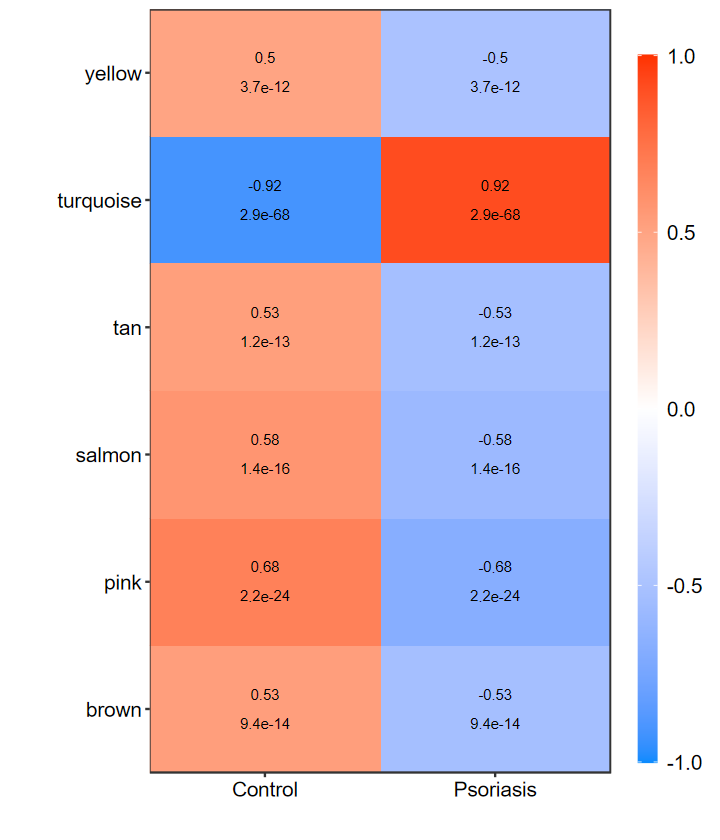
# 自定义显示的阈值
heatmap_replot(WGCNA_res$signif_summary, pvalue = 0.001, corr = 0.5)
get_hub_genes
- 待开发
- 根据相关性阈值和MM/GS阈值重新提取hub基因
- 目前根据返回的 WGCNA_res 对象有点R基础的也能自行提取
