R脚本--PCA分析系列1_v1.0
PCA分析系列-基于prcomp函数
目录
PCA分析系列-基于prcomp函数
摘要
1. 数据准备
2. R脚本
3. 结果示例
3.1 结果结构
3.2 原始矩阵展示
(1)gene_TPM2.txt
(2)group2.txt
3.3 图展示
(1)p_ggbiplot.png
(2)p_ggbiplot_lable.png
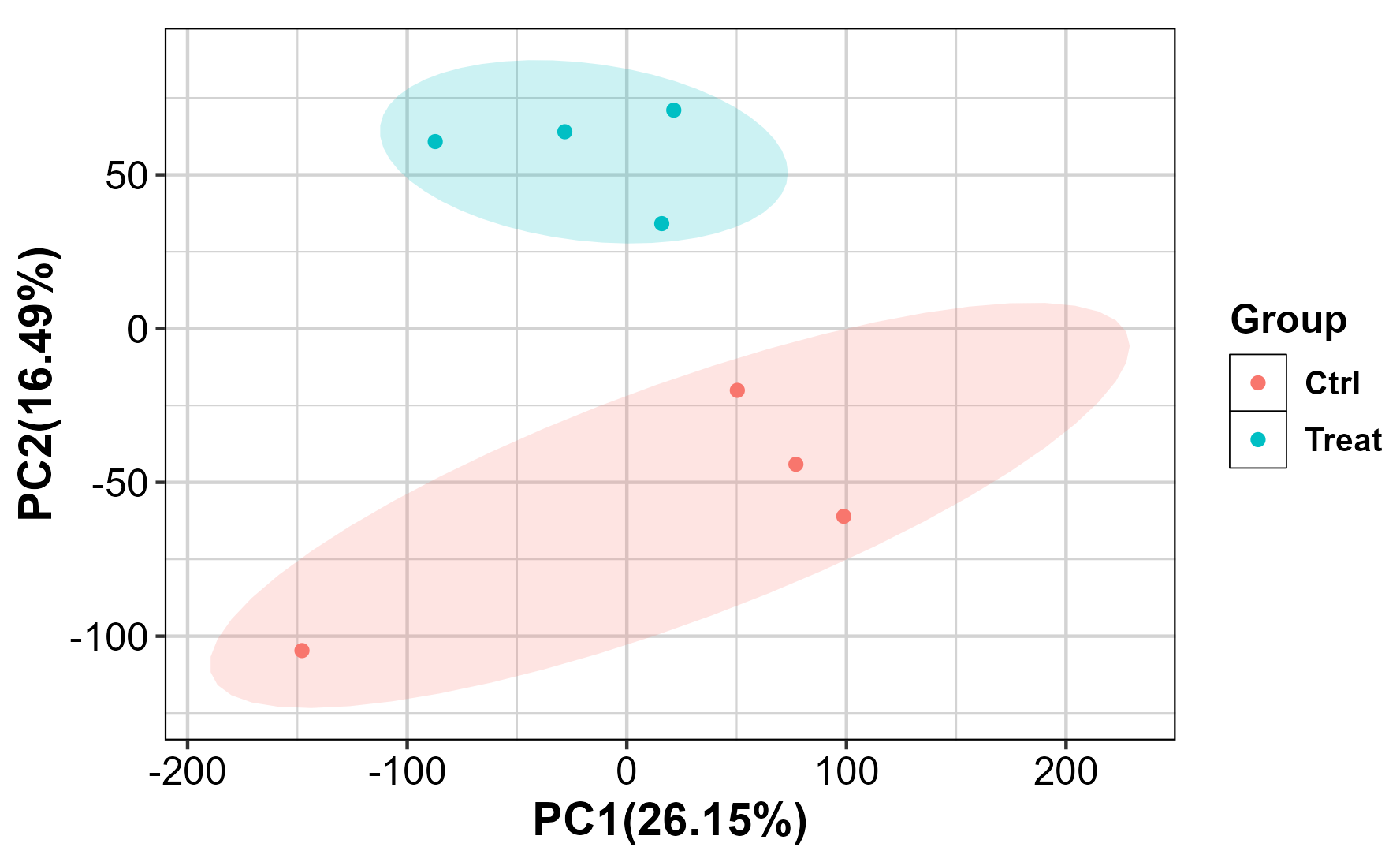
(3)p_ggplot2.png
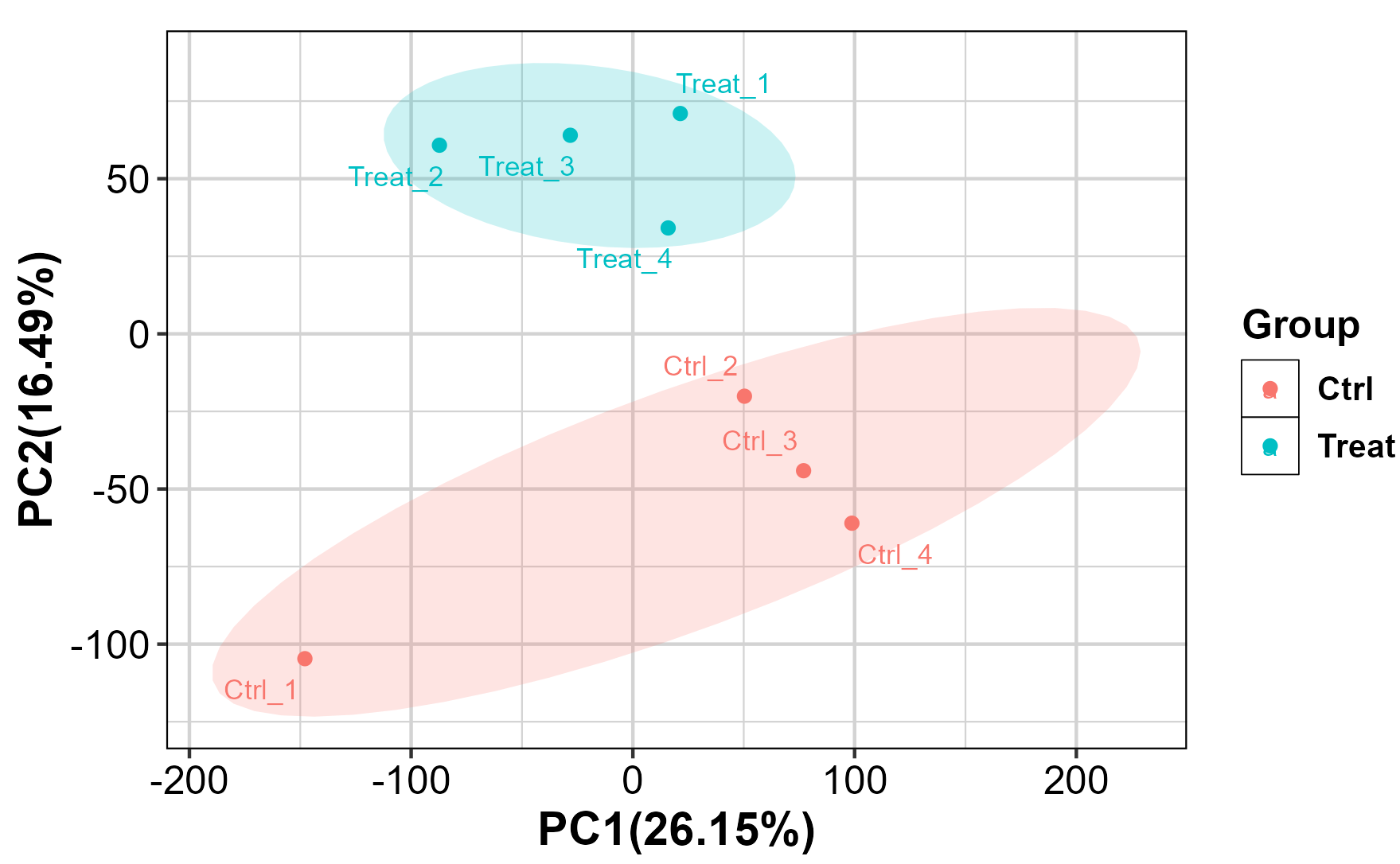
(4)p_ggplot2_lable.png
摘要
PCA分析R包有很多,不同的包作图时,如果不注意某些参数设置就会出现:画出图不一致情况。今天主要基于prcomp函数进行主成分分析,然后分别用ggbiplot包和ggplot2包进行可视化。注:如果要展示置信圈,要求每组组内样本数≥4;如果每组组内样本数=3,是画不了置信圈的,只能是画分组椭圆(一般也没必要,意义不大);如果每组组内样本数<3,尽量不要自找麻烦弄PCA了。
1. 数据准备
表达矩阵:第一列为基因信息,其余列为样本对应的表达数据
分组信息:第一列为样本,第二列为 分组
注:两个表样本要对称一致。
2. R脚本
rm(list = ls())# 加载所需包
library(openxlsx)
library(ggbiplot)
library(ggplot2)
library(ggrepel)
getwd()
# 设置工作目录
setwd("E:/SCRIPTS/PCA") #换成你实际的位置进行# 读取数据
##如果是xlsx文件
#dat <- read.xlsx("gene_TPM2.xlsx", colNames = TRUE, rowNames = TRUE)
#SampleDat <- read.table("group2.xlsx", colNames = TRUE, rowNames = TRUE)
dat <- read.table("gene_TPM2.txt", sep = '\t', row.names = 1, header = T, stringsAsFactors = F)
SampleDat <- read.table("group2.txt", sep = '\t', header = T)
#View(dat)
#View(SampleDat)
#View(SampleDat)# 数据处理
#datNew <- dat[, -1] # 前面加载数据时候设置row.names = 1,这里就不用
datNew <- dat
group <- as.character(SampleDat[match(colnames(datNew), SampleDat[, 1]), 2])#datNewT <- t(as.matrix(datNew))
#datNewT <- na.omit(datNewT)
#建议去掉常数列(全 0 或全相同值,全是0或相同值并不会提供变异值,对结果无贡献)
datNewT <- t(log2(as.matrix(dat) + 1)) # 先 log
datNewT <- datNewT[, apply(datNewT, 2, sd) > 0] # 再踢常数列# 计算PCA
#PCADat <- prcomp(datNewT)
#建议进行中心化和标准化处理,让所有基因处在“同一量尺”,使PCA不会被高表达基因绑架。
PCADat <- prcomp(datNewT, scale. = TRUE, center = TRUE)# 映射分组信息
df_pca <- as.data.frame(PCADat$x)
df_pca$group <- SampleDat$group
#View(df_pca)# 提取解释度
summ <- summary(PCADat)
#View(summ)
x_lable <- paste0("PC1(", round(summ$importance[2, 1]*100, 2), "%)")
y_lable <- paste0("PC2(", round(summ$importance[2, 2]*100, 2), "%)")# 创建保存图形的文件夹
if (!dir.exists("PCA_results/ggbiplot_method")) {dir.create("PCA_results/ggbiplot_method")
}
if (!dir.exists("PCA_results/ggplot2_method")) {dir.create("PCA_results/ggplot2_method")
}par(family = "Times New Roman") # 设置全局字体# 设置主题样式
theme_custom <- theme(plot.background = element_rect(fill = "white"), # 设置背景为白色panel.background = element_rect(fill = "white"), # 设置面板背景为白色panel.grid = element_line(color = "lightgray"), # 设置主要网格线颜色axis.text = element_text(color = "black", size = 12, face = "plain"), # 设置刻度值字体颜色、大小和粗细axis.title = element_text(color = "black", size = 14, face = "bold"), # 设置轴标签字体颜色、大小和斜体legend.text = element_text(color = "black", size = 10, face = "bold"), # 设置图例字体颜色、大小和样式legend.title = element_text(color = "black", size = 12, face = "bold"), # 设置图例标题字体颜色、大小和样式panel.border = element_rect(color = "black", fill = NA, linewidth = 0.5) # 设置面板边框为黑色
)################################# 无置信圈 #####################################
# -------------------- ggbiplot 图形 --------------------
# 基础图形
p_ggbiplot <- ggbiplot(PCADat, obs.scale = 1, var.scale = 0, ellipse = TRUE, circle = FALSE, var.axes = F, groups = group) +theme_custom + coord_fixed(ratio = 1.4) #调整x轴和y轴长度比例
print(p_ggbiplot)# 保存图形
ggsave("PCA_results/ggbiplot_method/p_ggbiplot.png", plot = p_ggbiplot, width = 6, height = 6, dpi = 300)
ggsave("PCA_results/ggbiplot_method/p_ggbiplot.pdf", plot = p_ggbiplot, width = 6, height = 6)dev.off()# 添加标签的图形
p_ggbiplot_lable <- p_ggbiplot + geom_text(aes(label = rownames(datNewT)), vjust = 1.5, size = 3, color = "black") + coord_fixed(ratio = 1.4) #调整x轴和y轴长度比例
print(p_ggbiplot_lable)ggsave("PCA_results/ggbiplot_method/p_ggbiplot_lable.png", plot = p_ggbiplot_lable, width = 6, height = 6, dpi = 300)
ggsave("PCA_results/ggbiplot_method/p_ggbiplot_lable.pdf", plot = p_ggbiplot_lable, width = 6, height = 6)dev.off()################################# 有置信圈 #####################################
# -------------------- ggplot2 图形 --------------------
# 基础图形
p_ggplot2 <- ggplot(df_pca, aes(x = PC1, y = PC2,colour = group)) + stat_ellipse(aes(fill = group),type = "norm", geom = "polygon", alpha = 0.2, level = 0.68,color = NA, linetype = 2, show.legend = FALSE) + geom_point() + labs(x = x_lable, y = y_lable, color = "Group") + guides(fill = "none") + theme(legend.key = element_rect(colour = "black", # 黑色边框size = 0.2 # 边框线宽)) + theme_custom + coord_fixed(ratio = 1.4) #调整x轴和y轴长度比例print(p_ggplot2)# 保存图形
ggsave("PCA_results/ggplot2_method/p_ggplot2.png", plot = p_ggplot2, width = 6, height = 6, dpi = 300)
ggsave("PCA_results/ggplot2_method/p_ggplot2.pdf", plot = p_ggplot2, width = 6, height = 6)dev.off()# 添加标签的图形
p_ggplot2_lable <- ggplot(df_pca,aes(x = PC1,y = PC2,colour = group)) +stat_ellipse(aes(fill = group),type = "norm",geom = "polygon",alpha = 0.2,color = NA,level = 0.68, linetype = 2,show.legend = FALSE) +geom_point() +geom_text_repel(aes(label = rownames(df_pca)),size = 3,box.padding = 0.35,point.padding = 0.5,segment.color = "grey50") +labs(x = x_lable,y = y_lable,color = "Group") +theme(legend.key = element_rect(colour = "black", size = 0.2)) + guides(fill = "none") + theme_custom + coord_fixed(ratio = 1.4) #调整x轴和y轴长度比例print(p_ggplot2_lable)ggsave("PCA_results/ggplot2_method/p_ggplot2_lable.png", plot = p_ggplot2_lable, width = 6, height = 6, dpi = 300)
ggsave("PCA_results/ggplot2_method/p_ggplot2_lable.pdf", plot = p_ggplot2_lable, width = 6, height = 6)# 关闭图形设备(根据实际需求选择是否保留)
dev.off()
3. 结果示例
3.1 结果结构
当前工作路径/
├── PCA_results/
│ ├── ggbiplot_method/
│ │ ├── p_ggbiplot.png
│ │ ├── p_ggbiplot.pdf
│ │ ├── p_ggbiplot_lable.png
│ │ └── p_ggbiplot_lable.pdf
│ └── ggplot2_method/
│ ├── p_ggplot2.png
│ ├── p_ggplot2.pdf
│ ├── p_ggplot2_lable.png
│ └── p_ggplot2_lable.pdf
├── gene_TPM2.txt
├── group2.txt
└── PCA_script.R
3.2 原始矩阵展示
(1)gene_TPM2.txt
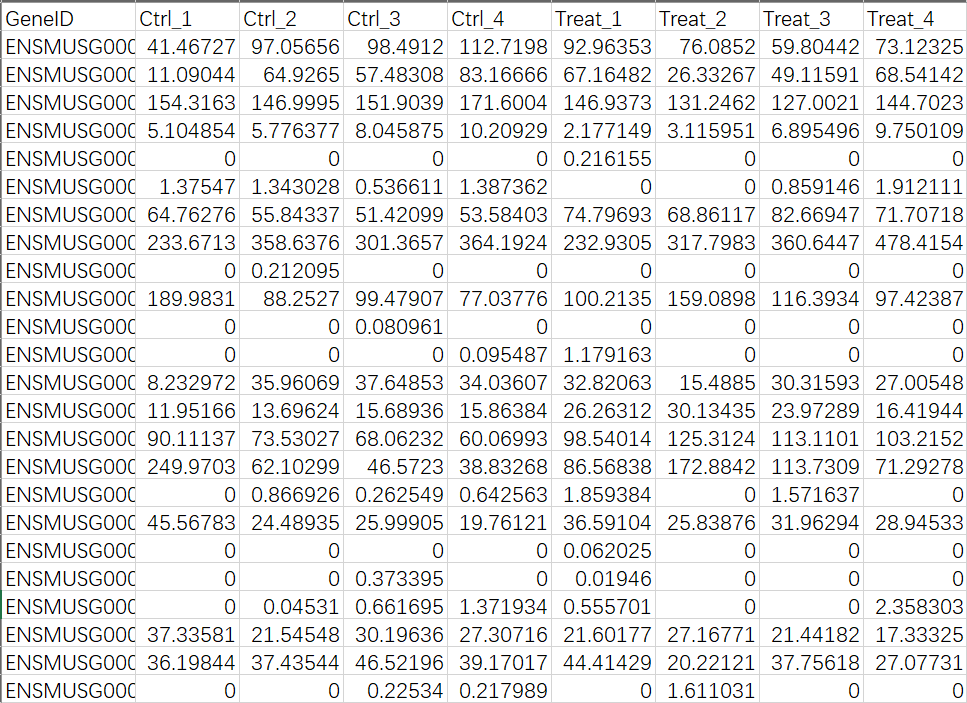
注:示例数据为随机生成结果,并没有从原始count计数矩阵进行过滤和转换等操作,所以会有部分基因表达很异常。
(2)group2.txt
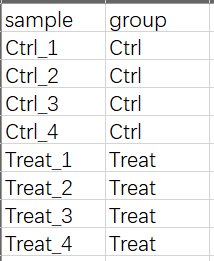
注:额外说一下,就是txt文件是可以使用表格工具打开查看的,生信分析中.txt/.xlsx等只是存储数据的格式而已,重要的是存储时数据分割形式,后缀意义并不大(仅限于像这种矩阵哈)。
3.3 图展示
(1)p_ggbiplot.png
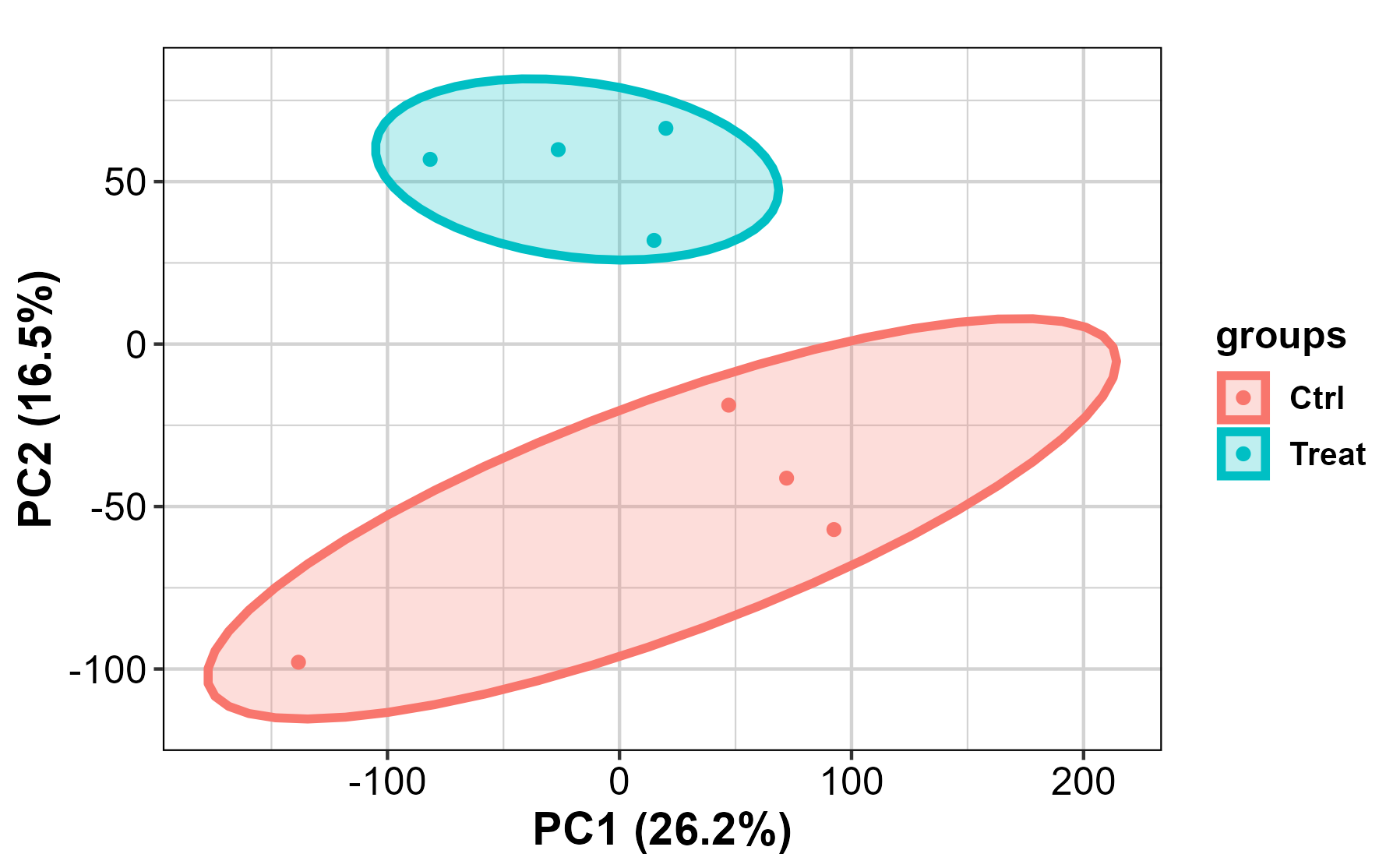
(2)p_ggbiplot_lable.png
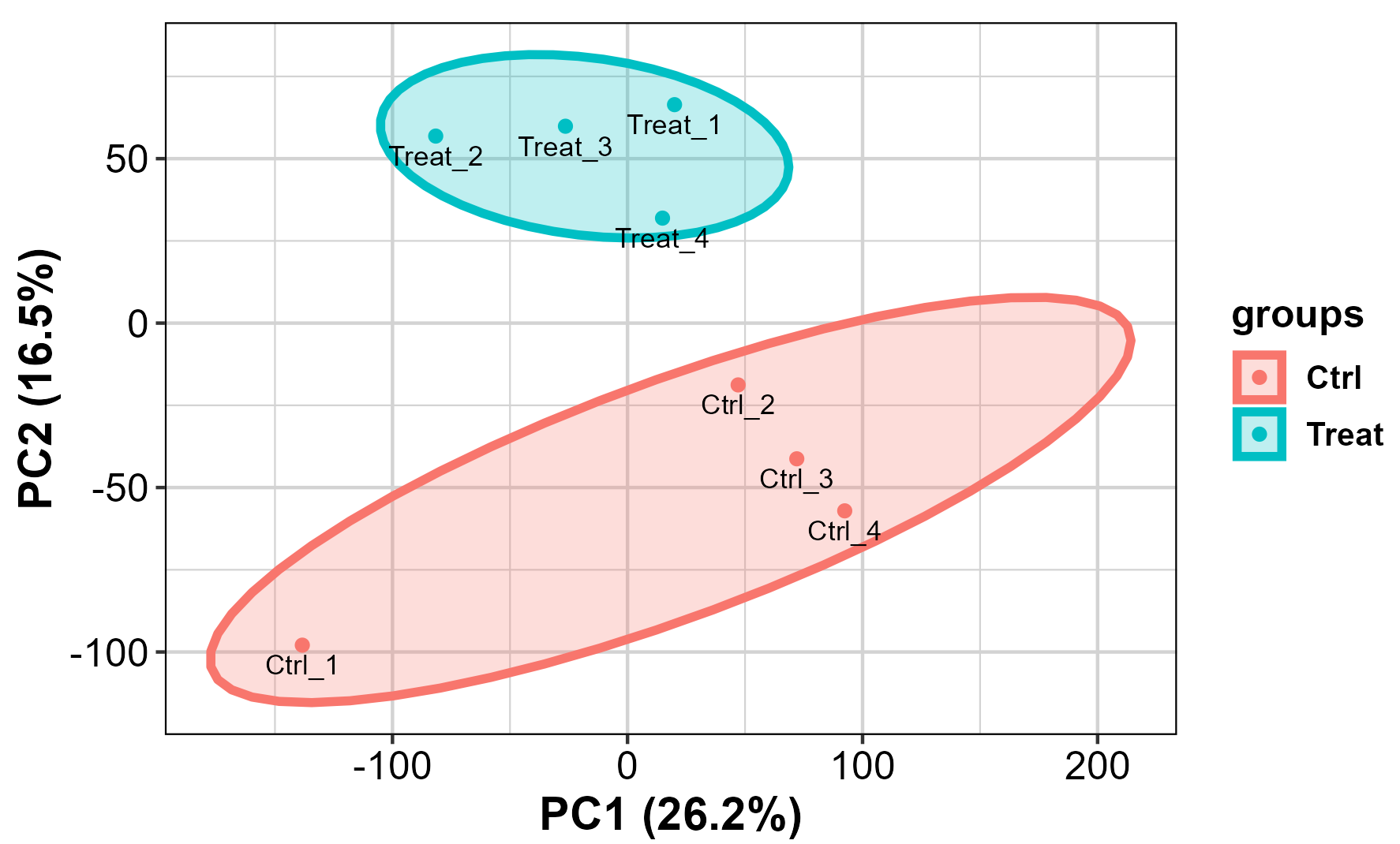
(3)p_ggplot2.png
(4)p_ggplot2_lable.png
写作不易,要是有补充的请call me,相互学习促进,谢谢!
