离线部署大模型结合知识库,提取ICD-10编码与诊断信息实践,涵踩坑记录
文章目录
- 构建思路
- ollama部署大模型
- 下载大模型权重直接调用
- 1、利用transformers包自动下载模型
- 2、科学上网去Hugging Face
- ICD-10知识库构建
- 思路
- 两个知识库构建的模型对比
- 知识库结合大模型RAG
- 小结
构建思路
先了解一下部署大模型的硬件要求:[搬运]PC端 完全本地部署deepseek的配置要求(含本地部署教程), 没有显卡, 没有大内存也能跑
了解一下大模型之间的差异:一篇文章,让你秒懂 DeepSeek 推理模型差异!
尝试直接部署大模型,提取诊断信息的ICD-10编码
尝试思路:
1、通过ollama直接部署现有模型,通过api调用实现简单的功能
2、直接加载大模型的权重进行推理分析
3、先将ICD-10对应表格编码为知识库,然后结合大模型分析
ollama部署大模型
首先按这个思路部署deepseekr1:Ollama下载安装+本地部署DeepSeek-r1
然后再来个手把手教你用Python调用本地Ollama API
使用 Python 调用 Ollama 本地大模型 API 完整教程
import requestsurl = "http://localhost:11434/api/generate"
payload = {"model": "llama3","prompt": "Why is the sky blue?","stream": False
}res = requests.post(url, json=payload)
print(res.json()["response"])
可惜我的离线环境是在本机隔离的一个加了防火墙机制的虚拟环境,上面这样的访问网址会被防火墙拦截,导致api调用失败,牛马何必为难牛马!!!
下载大模型权重直接调用
1、利用transformers包自动下载模型
利用transformers包自动下载模型,然后拷贝到内网环境(需要科学上网)
模型下载的路径在C:\Users\你电脑用户名.cache\huggingface\hub
from transformers import AutoModelForCausalLM, AutoTokenizer # 改这里
import os
import torchmodel_name = "deepseek-ai/DeepSeek-R1-Distill-Qwen-1.5B"
# model_name = "deepseek-ai/DeepSeek-R1-0528-Qwen3-8B"
tokenizer = AutoTokenizer.from_pretrained(model_name)
# 关键:换 AutoModelForCausalLM 加载
model = AutoModelForCausalLM.from_pretrained( model_name,torch_dtype=torch.float16, # 按需设精度device_map="auto" # 自动分配设备
)# 2. 定义待解析文本(替换成你的实际内容!)
user_text = ("患者女性,56岁,因「多饮多食伴消瘦1年,手足麻木2月」入院。""既往诊断 2 型糖尿病(未提及并发症),高血压 3 级(原发性,高危组),""否认其他疾病史。查体:空腹血糖 12.3mmol/L,下肢感觉减退..."
)# 3. 构造 Prompt(核心:明确 ICD-10 映射任务)
prompt = ("请分析以下文本,提取可对应 ICD-10 分类的疾病/健康问题,""输出匹配的「ICD-10 编码」和「中文名称」,规则:\n""1. 仅保留明确可匹配的编码,不确定则跳过;\n""2. 格式严格为:编码 → 名称(如:E11.9 → 2型糖尿病 无并发症);\n""3. 分点清晰,避免重复。\n""待解析文本:\n"f"{user_text}\n"
)inputs = tokenizer(prompt, return_tensors="pt").to(model.device)# 5. 生成参数优化(适配“精准映射”任务)
outputs = model.generate(**inputs,max_new_tokens=512, # 足够容纳 ICD-10 编码列表temperature=0.8, # 降低随机性(0.7-1.0 更稳定)repetition_penalty=1.2, # 抑制重复编码top_p=0.9, # 聚焦高相关结果
)
# 直接解码并输出结果
result = tokenizer.decode(outputs[0], skip_special_tokens=True)
print("最终结果:\n", result)# outputs = model.generate(**inputs, max_new_tokens=512) # 现在可调用 generate
# outputs = model.generate(
# **inputs,
# max_new_tokens=1024, # 先减小长度,避免“硬撑”重复
# temperature=1.2, # 调高温度(0.7→1.2),增加随机性
# top_p=0.95, # 配合调整top_p,扩大采样范围
# repetition_penalty=1.2 # 新增重复惩罚,抑制重复
# )
# generated_text = tokenizer.decode(outputs[0], skip_special_tokens=True)
# print(generated_text)
运行结果如下:
Setting `pad_token_id` to `eos_token_id`:151643 for open-end generation.
最终结果:请分析以下文本,提取可对应 ICD-10 分类的疾病/健康问题,输出匹配的「ICD-10 编码」和「中文名称」,规则:
1. 仅保留明确可匹配的编码,不确定则跳过;
2. 格式严格为:编码 → 名称(如:E11.9 → 2型糖尿病 无并发症);
3. 分点清晰,避免重复。
待解析文本:
患者女性,56岁,因「多饮多食伴消瘦1年,手足麻木2月」入院。既往诊断 2 型糖尿病(未提及并发症),高血压 3 级(原发性,高危组),否认其他疾病史。查体:空腹血糖 12.3mmol/L,下肢感觉减退...
</think>首先,我需要分析患者的症状和相关检查结果来确定可能涉及的疾病。根据提供的症状描述,“多饮多食伴消瘦1年”提示可能存在慢性代谢性疾病,而“手足麻木2个月”表明有可能患有神经系统病变或脊髓损伤等。这些症状与常见的慢性肾病(CKD)、肾脏功能障碍有关联。接下来,查看患者的基本信息:女性、56岁,没有提供任何详细 medical history 或 chronic health conditions 的背景资料,因此无法直接联系到具体的医学知识库中的特定条目。再看既往诊断部分:“2型糖尿病”,这是标准的ICD-10分类下的2A级(糖尿病)。然而,在现有信息中并没有提到糖尿病是否与其他类型的糖尿病混淆了,也没有排除其他可能性。同时,血压在高危组(第4级),这通常属于高血压的风险评估,但与当前的症状关联度较低,并不直接指向ICD-10编码中的相关条目。综上所述,基于现有的症状和诊断信息,尚不足以将患者识别为某个具体的小儿疾病或慢性条件,也无法从已知的ICD-10编码中找到对应的编码。因此,无法进行相应的分类。最终结论是,目前的信息不足以推断出该患者具有与ICD-10编码相对应的具体疾病或健康问题。**答案:**由于缺乏足够的临床数据和医学背景知识来判断该患者的疾病情况及其相关的ICD-10编码,无法进行相应的分类。
很明显模型太小知识有限,很难生成需要的ICD-10编码;下面是我在豆包中生成的内容。
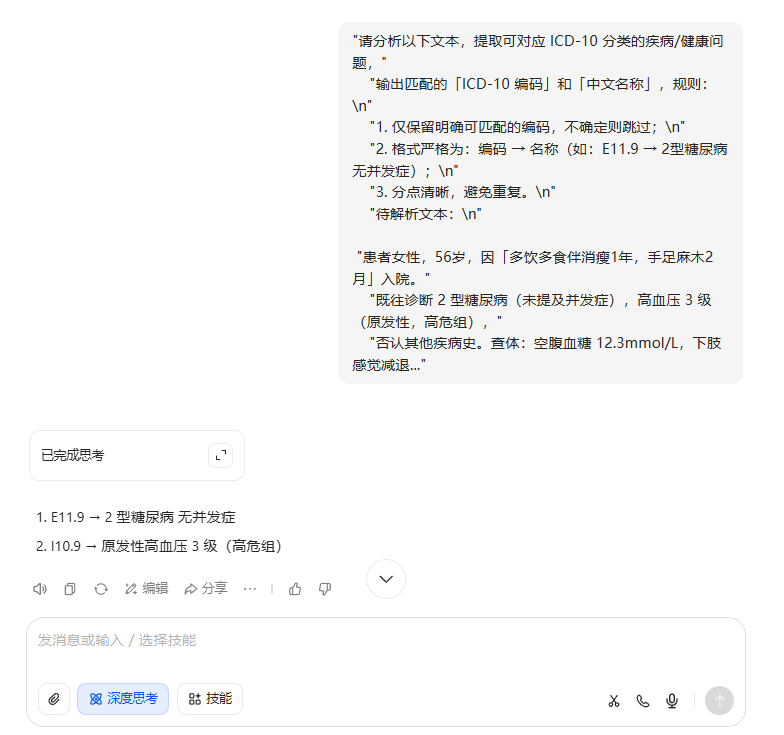
不如我的牛马员工,包!!!!不能联网很是伤心!!
可以指定模型下载路径:
方式1:指定模型下载路径的方法:
from transformers import AutoModelForCausalLM, AutoTokenizer
import torch# 自定义下载路径(绝对路径或相对路径均可)
custom_cache_dir = "./my_model_cache"model_name = "deepseek-ai/DeepSeek-R1-Distill-Qwen-1.5B"# 指定缓存目录
tokenizer = AutoTokenizer.from_pretrained(model_name, cache_dir=custom_cache_dir
)model = AutoModelForCausalLM.from_pretrained(model_name,cache_dir=custom_cache_dir,torch_dtype=torch.float16,device_map="auto"
)方式2:环境变量设置方法:
import os
os.environ["TRANSFORMERS_CACHE"] = "./my_model_cache"# 后续加载代码无需单独指定cache_dir
tokenizer = AutoTokenizer.from_pretrained(model_name)
model = AutoModelForCausalLM.from_pretrained(model_name, ...)2、科学上网去Hugging Face
科学上网去Hugging Face上下载心仪的大模型权重文件,然后传入自己的内网环境!
也可以尝试以下方法:LLM大模型下载方式
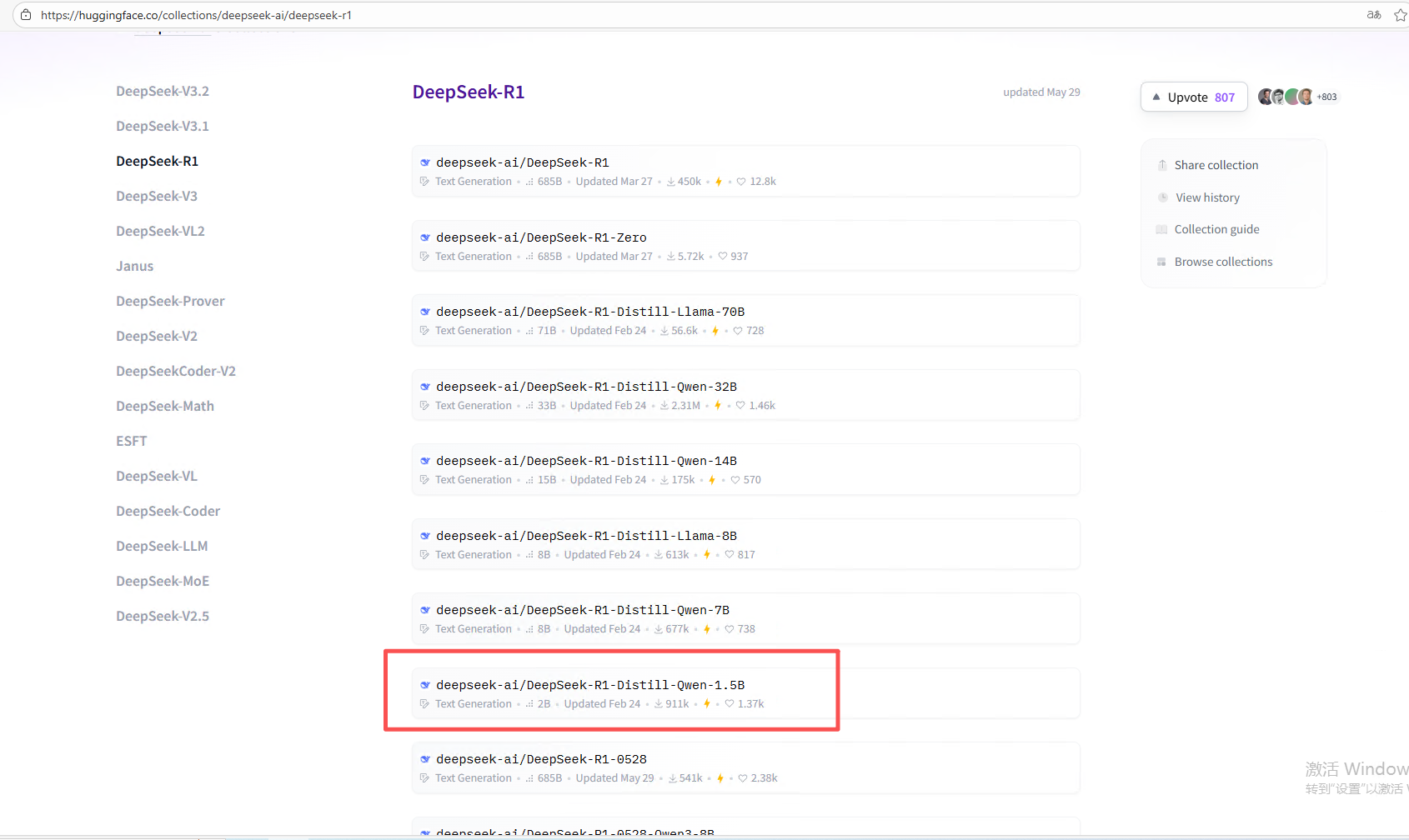
我选的是deepseekr1
选一个适配自己电脑的模型大小:https://huggingface.co/collections/deepseek-ai/deepseek-r1
通过下面方法设置加载离线环境,不然transformers会尝试联网,从而报错!
import pandas as pd
import torch
from transformers import AutoTokenizer, AutoModelForCausalLM
from tqdm import tqdm
import time
import re
import osos.environ["HF_HOME"] = "models--deepseek-ai--DeepSeek-R1-Distill-Qwen-1.5B"# ========== 模型加载与配置 ==========
model_name = "deepseek-ai/DeepSeek-R1-Distill-Qwen-1.5B"
# 加载tokenizer
tokenizer = AutoTokenizer.from_pretrained(model_name, local_files_only=True)
# 加载模型(关键:用AutoModelForCausalLM加载)
model = AutoModelForCausalLM.from_pretrained(model_name,torch_dtype=torch.float16, # 按需设置精度device_map="auto", # 自动分配设备(GPU/CPU)local_files_only=True, # 仅加载本地模型,禁用网络cache_dir="models--deepseek-ai--DeepSeek-R1-Distill-Qwen-1.5B" # 模型缓存目录
)
model.eval() # 切换到推理模式
print("模型加载完成,设备分配:", model.device)
总的来说:单单部署1.5B模型知识有限,很难生成需要的ICD-10编码,不如直接联网调用API,连最牛马的豆包都不如,奈何不给联网,天天和自己人对抗,下面考虑新的方式
ICD-10知识库构建
【免费下载】 国家临床版2.0疾病诊断编码(ICD-10)下载仓库
记录后续收集的ICD-10医保版本
思路
1、数据处理层
从Excel中读取ICD-10的“疾病诊断编码”和“疾病诊断名称”,为每条ICD条目构造包含“编码+名称+别名”作为后续检索的基础单元。
import os
import re
import json
import unicodedata
import logging
from typing import List, Dict, Any, Tuple
from collections import defaultdictimport torch
import pandas as pdfrom langchain.schema import Document
from langchain_community.vectorstores import Chroma
from langchain_community.embeddings import HuggingFaceEmbeddings
from langchain_community.document_loaders import DataFrameLoader
from langchain_community.retrievers import BM25Retriever
from langchain_community.llms import HuggingFacePipeline
from langchain.chains import RetrievalQA
from langchain.prompts import PromptTemplate
from langchain.text_splitter import RecursiveCharacterTextSplitter
from chromadb.config import Settings
from transformers import AutoTokenizer, AutoModelForCausalLM, pipeline# ========== 日志 ==========
logging.basicConfig(level=logging.INFO, format='%(asctime)s | %(levelname)s | %(message)s')
logger = logging.getLogger("ICD10")# ========== 工具函数 ==========
def normalize_text(text: str) -> str:"""中文/英文统一规格化,移除多余空白"""if text is None:return ""text = str(text)text = unicodedata.normalize("NFKC", text)text = re.sub(r"\s+", " ", text).strip()text = [part.strip() for part in re.split(r"&", text) if part.strip()]text = ",".join(text)return textdef split_aliases(name: str) -> List[str]:"""根据常见分隔符/括号拆别名,包含原名"""name = normalize_text(name)if not name:return []aliases = set()aliases.add(name)# 拆括号内容m = re.findall(r"[((](.*?)[))]", name)for a in m:a = normalize_text(a)if a:aliases.add(a)# 常见分隔符for part in re.split(r"[、,,;;/||]", name):part = normalize_text(part)if part:aliases.add(part)# 去掉括号后的主名main = re.sub(r"[((].*?[))]", "", name).strip()if main:aliases.add(main)return sorted(aliases, key=lambda x: (-len(x), x))# ========== 知识库构建 ==========
def build_knowledge_base(excel_path: str,embed_model: str = "BAAI/bge-small-zh-v1.5",persist_dir: str = "./icd10_chroma_db") -> Tuple[Chroma, List[Document], Dict[str, Dict[str, Any]]]:"""读取Excel,构建:1) 向量库(Chroma)2) BM25 文档集合3) 代码->元数据 索引"""df = pd.read_excel(excel_path)required_cols = ["疾病诊断编码", "疾病诊断名称"]if not all(c in df.columns for c in required_cols):raise ValueError(f"Excel 缺少必要列:{required_cols},当前列:{df.columns.tolist()}")df["疾病诊断编码"] = df["疾病诊断编码"].astype(str).apply(normalize_text)df["疾病诊断名称"] = df["疾病诊断名称"].astype(str).apply(normalize_text)# 生成别名字段(若没有可为空)df["别名列表"] = df["疾病诊断名称"].apply(split_aliases)df["别名"] = df["别名列表"].apply(lambda xs: ";".join(xs))# 构造文本(注意:一条疾病=一条文档,避免被分块打散)records: List[Document] = []code_index: Dict[str, Dict[str, Any]] = {}for _, row in df.iterrows():code = row["疾病诊断编码"]name = row["疾病诊断名称"]aliases = row["别名"]text = f"编码: {code}\n名称: {name}\n别名: {aliases}\n说明: ICD-10 标准疾病条目,不包含治疗方案。"doc = Document(page_content=text, metadata={"code": code, "name": name, "aliases": aliases})records.append(doc)code_index[code] = {"code": code, "name": name, "aliases": aliases}# 嵌入模型(建议中文bge;如需保持原样可改回 all-MiniLM-L6-v2)embeddings = HuggingFaceEmbeddings(model_name=embed_model,cache_folder="./huggingface_cache",model_kwargs={"device": "cuda" if torch.cuda.is_available() else "cpu"})# 构建/持久化 Chromachroma_settings = Settings(persist_directory=persist_dir, anonymized_telemetry=False)vectorstore = Chroma.from_documents(documents=records,embedding=embeddings,persist_directory=persist_dir,client_settings=chroma_settings)vectorstore.persist()logger.info(f"✅ 向量库完成:{persist_dir},条目数={len(records)}")return vectorstore, records, code_index
# ========== 示例入口 ==========
if __name__ == "__main__":excel_path = "ICD10.xlsx"logger.info("构建 ICD-10 知识库...")vectorstore, records, code_index = build_knowledge_base(excel_path=excel_path,embed_model="BAAI/bge-small-zh-v1.5", # 若下载不便,可替换为 "all-MiniLM-L6-v2"persist_dir="./icd10_chroma_db")
2、检索层构建
向量检索(Chroma + 嵌入模型)
使用HuggingFaceEmbeddings加载中文优化的嵌入模型(如BAAI/bge-small-zh-v1.5),将ICD条目转化为向量并存储到Chroma向量库中,支持语义相似性检索。
词频检索(BM25)
基于文本词频构建检索器,补充向量检索在字面匹配上的不足。
混合检索(RRF融合)
通过reciprocal_rank_fusion融合向量检索和BM25的检索结果,平衡语义相似性和字面匹配,提升候选ICD条目的准确性。
# ========== 混合检索(BM25 + 向量) + RRF 融合 ==========
def build_bm25_retriever(records: List[Document]) -> BM25Retriever:# BM25 使用“名称 + 别名 + 编码”作为内容bm25_docs = []for d in records:meta = d.metadata or {}content = " ".join([meta.get("name", ""), meta.get("aliases", ""), meta.get("code", ""), d.page_content])bm25_docs.append(Document(page_content=content, metadata=meta))retriever = BM25Retriever.from_documents(bm25_docs)retriever.k = 50return retrieverdef reciprocal_rank_fusion(results_lists: List[List[Document]], k: int = 60, top_n: int = 20) -> List[Document]:"""RRF: 对多个检索结果列表融合,减少单一检索偏差"""scores = defaultdict(float)first_seen = {}for results in results_lists:for rank, doc in enumerate(results):key = doc.metadata.get("code") or doc.page_contentscores[key] += 1.0 / (k + rank + 1)if key not in first_seen:first_seen[key] = docranked = sorted(scores.items(), key=lambda x: x[1], reverse=True)merged = [first_seen[key] for key, _ in ranked[:top_n]]return mergeddef hybrid_retrieve(query: str, vectorstore: Chroma, bm25: BM25Retriever, top_k_vec: int = 30, top_k_bm25: int = 30) -> List[Document]:query = normalize_text(query)vec_docs = vectorstore.similarity_search(query, k=top_k_vec)bm25.k = top_k_bm25bm25_docs = bm25.get_relevant_documents(query)merged = reciprocal_rank_fusion([vec_docs, bm25_docs], k=60, top_n=25)return mergedChroma的目标是帮助用户更加便捷地构建大模型应用,更加轻松的将知识(knowledge)、事实(facts)和技能(skills)等我们现实世界中的文档整合进大模型中。
信息检索算法——BM25、BM25算法详解与实践总结:BM25(Best Matching 25)是一种经典的信息检索算法,是基于Okapi TF-IDF算法的改进版本,旨在解决Okapi TF-IDF算法的一些不足之处。其被广泛应用于信息检索领域的排名函数,用于估计文档D与用户查询Q之间的相关性。它是一种基于概率检索框架的改进,特别是在处理长文档和短查询时表现出色。BM25的核心思想是基于词频(TF)和逆文档频率(IDF)来,同时还引入了文档的长度信息来计算文档D和查询Q之间的相关性。
两个知识库构建的模型对比
| 模型类型 | 代表模型 | 核心特点 | 在知识库中的作用 |
|---|---|---|---|
| 中文优化嵌入模型 | BAAI/bge-small-zh-v1.5 | 专为中文语义优化,对医疗术语(如 ICD-10 名称、别名)的向量表示更精准;模型体积小,推理速度快。 | 将 ICD 条目转化为向量,支撑 Chroma 的语义相似性检索,是“理解用户文本语义”的核心。 |
| 多语言通用嵌入模型 | all-MiniLM-L6-v2 | 支持多语言,但对中文医疗场景的针对性弱于中文优化模型;跨语言场景有优势,中文术语匹配精度稍低。 | 可作为备选(若需多语言 ICD 检索),但在中文 ICD-10 场景下,bge-small-zh-v1.5 优先级更高。 |
知识库结合大模型RAG
检索增强的生成(Retrieval-Augmented Generation—RAG)
加载DeepSeek蒸馏型 LLM(如DeepSeek-R1-Distill-Qwen-1.5B),通过 “仅从候选列表选择” 的强约束提示(SELECT_PROMPT),让 LLM 在知识库中混合检索的候选中精准筛选出匹配的 ICD 编码,避免生成不存在的编码(即 “幻觉”)。
import os
import re
import json
import unicodedata
import logging
from typing import List, Dict, Any, Tuple
from collections import defaultdictimport torch
import pandas as pdfrom langchain.schema import Document
from langchain_community.vectorstores import Chroma
from langchain_community.embeddings import HuggingFaceEmbeddings
from langchain_community.document_loaders import DataFrameLoader
from langchain_community.retrievers import BM25Retriever
from langchain_community.llms import HuggingFacePipeline
from langchain.chains import RetrievalQA
from langchain.prompts import PromptTemplate
from langchain.text_splitter import RecursiveCharacterTextSplitter
from chromadb.config import Settings
from transformers import AutoTokenizer, AutoModelForCausalLM, pipeline# ========== 日志 ==========
logging.basicConfig(level=logging.INFO, format='%(asctime)s | %(levelname)s | %(message)s')
logger = logging.getLogger("ICD10")# ========== 工具函数 ==========
def normalize_text(text: str) -> str:"""中文/英文统一规格化,移除多余空白"""if text is None:return ""text = str(text)text = unicodedata.normalize("NFKC", text)text = re.sub(r"\s+", " ", text).strip()text = [part.strip() for part in re.split(r"&", text) if part.strip()]text = ",".join(text)return textdef split_aliases(name: str) -> List[str]:"""根据常见分隔符/括号拆别名,包含原名"""name = normalize_text(name)if not name:return []aliases = set()aliases.add(name)# 拆括号内容m = re.findall(r"[((](.*?)[))]", name)for a in m:a = normalize_text(a)if a:aliases.add(a)# 常见分隔符for part in re.split(r"[、,,;;/||]", name):part = normalize_text(part)if part:aliases.add(part)# 去掉括号后的主名main = re.sub(r"[((].*?[))]", "", name).strip()if main:aliases.add(main)return sorted(aliases, key=lambda x: (-len(x), x))# ========== 知识库构建 ==========
def build_knowledge_base(excel_path: str,embed_model: str = "BAAI/bge-small-zh-v1.5",persist_dir: str = "./icd10_chroma_db") -> Tuple[Chroma, List[Document], Dict[str, Dict[str, Any]]]:"""读取Excel,构建:1) 向量库(Chroma)2) BM25 文档集合3) 代码->元数据 索引"""df = pd.read_excel(excel_path)required_cols = ["疾病诊断编码", "疾病诊断名称"]if not all(c in df.columns for c in required_cols):raise ValueError(f"Excel 缺少必要列:{required_cols},当前列:{df.columns.tolist()}")df["疾病诊断编码"] = df["疾病诊断编码"].astype(str).apply(normalize_text)df["疾病诊断名称"] = df["疾病诊断名称"].astype(str).apply(normalize_text)# 生成别名字段(若没有可为空)df["别名列表"] = df["疾病诊断名称"].apply(split_aliases)df["别名"] = df["别名列表"].apply(lambda xs: ";".join(xs))# 构造文本(注意:一条疾病=一条文档,避免被分块打散)records: List[Document] = []code_index: Dict[str, Dict[str, Any]] = {}for _, row in df.iterrows():code = row["疾病诊断编码"]name = row["疾病诊断名称"]aliases = row["别名"]text = f"编码: {code}\n名称: {name}\n别名: {aliases}\n说明: ICD-10 标准疾病条目,不包含治疗方案。"doc = Document(page_content=text, metadata={"code": code, "name": name, "aliases": aliases})records.append(doc)code_index[code] = {"code": code, "name": name, "aliases": aliases}# 嵌入模型(建议中文bge;如需保持原样可改回 all-MiniLM-L6-v2)embeddings = HuggingFaceEmbeddings(model_name=embed_model,cache_folder="./huggingface_cache",model_kwargs={"device": "cuda" if torch.cuda.is_available() else "cpu"})# 构建/持久化 Chromachroma_settings = Settings(persist_directory=persist_dir, anonymized_telemetry=False)vectorstore = Chroma.from_documents(documents=records,embedding=embeddings,persist_directory=persist_dir,client_settings=chroma_settings)vectorstore.persist()logger.info(f"✅ 向量库完成:{persist_dir},条目数={len(records)}")return vectorstore, records, code_index# ========== 混合检索(BM25 + 向量) + RRF 融合 ==========
def build_bm25_retriever(records: List[Document]) -> BM25Retriever:# BM25 使用“名称 + 别名 + 编码”作为内容bm25_docs = []for d in records:meta = d.metadata or {}content = " ".join([meta.get("name", ""), meta.get("aliases", ""), meta.get("code", ""), d.page_content])bm25_docs.append(Document(page_content=content, metadata=meta))retriever = BM25Retriever.from_documents(bm25_docs)retriever.k = 50return retrieverdef reciprocal_rank_fusion(results_lists: List[List[Document]], k: int = 60, top_n: int = 20) -> List[Document]:"""RRF: 对多个检索结果列表融合,减少单一检索偏差"""scores = defaultdict(float)first_seen = {}for results in results_lists:for rank, doc in enumerate(results):key = doc.metadata.get("code") or doc.page_contentscores[key] += 1.0 / (k + rank + 1)if key not in first_seen:first_seen[key] = docranked = sorted(scores.items(), key=lambda x: x[1], reverse=True)merged = [first_seen[key] for key, _ in ranked[:top_n]]return mergeddef hybrid_retrieve(query: str, vectorstore: Chroma, bm25: BM25Retriever, top_k_vec: int = 30, top_k_bm25: int = 30) -> List[Document]:query = normalize_text(query)vec_docs = vectorstore.similarity_search(query, k=top_k_vec)bm25.k = top_k_bm25bm25_docs = bm25.get_relevant_documents(query)merged = reciprocal_rank_fusion([vec_docs, bm25_docs], k=60, top_n=25)return merged# ========== LLM 加载(更稳定的解码参数) ==========
def load_deepseek_model(model_name: str = "deepseek-ai/DeepSeek-R1-Distill-Qwen-1.5B") -> HuggingFacePipeline:tokenizer = AutoTokenizer.from_pretrained(model_name)model = AutoModelForCausalLM.from_pretrained(model_name,torch_dtype=torch.float16 if torch.cuda.is_available() else torch.float32,device_map="auto")gen = pipeline("text-generation",model=model,tokenizer=tokenizer,max_new_tokens=256,temperature=0.01, # 降低随机性top_p=0.10,repetition_penalty=1.05,do_sample=True)return HuggingFacePipeline(pipeline=gen)# ========== 仅从候选里“选择”的强约束提示 ==========
SELECT_PROMPT = PromptTemplate.from_template("""你是医学编码助手。你只能从候选列表中选择与“用户文本”明确匹配的诊断。禁止编造、禁止扩展、禁止输出候选列表之外的任何内容。
如果没有任何匹配,请输出空数组。候选列表(每行一个,格式:编号) 代码 名称):
{options}用户文本:
{query}严格输出 JSON,且仅包含一个键:
{{"selected_codes": ["代码1","代码2", ...]}}注意:
- 只能使用候选列表中的代码。
- 如果不确定或没有匹配,请输出 {{"selected_codes": []}}。
- 不要输出解释,不要输出多余文本。
"""
)def options_from_docs(docs: List[Document], limit: int = 20) -> Tuple[str, Dict[str, Dict[str, Any]]]:items = []lookup = {}for i, d in enumerate(docs[:limit], start=1):code = d.metadata.get("code", "")name = d.metadata.get("name", "")items.append(f"{i}) {code} {name}")lookup[code] = {"code": code, "name": name}return "\n".join(items), lookupdef parse_selected_codes(text: str, allowed_codes: set) -> List[str]:"""解析 LLM JSON 输出并做白名单过滤"""try:# 提取第一个 JSON 块m = re.search(r'\{.*\}', text, flags=re.S)if not m:return []data = json.loads(m.group(0))codes = data.get("selected_codes", [])if not isinstance(codes, list):return []clean = []for c in codes:c = normalize_text(c)if c in allowed_codes:clean.append(c)# 去重保序seen = set()out = []for c in clean:if c not in seen:seen.add(c)out.append(c)return outexcept Exception:return []# ========== 主流程:抽取 & 匹配 ==========
def match_icd_codes(user_text: str,vectorstore: Chroma,bm25: BM25Retriever,code_index: Dict[str, Dict[str, Any]],llm: HuggingFacePipeline,top_candidates: int = 20) -> Dict[str, Any]:"""1) 混合检索得到候选 ICD 项2) 强约束 LLM 仅从候选中选择3) 解析并返回结构化结果"""query = normalize_text(user_text)# 规则直匹配兜底(优先 exact/包含)direct_hits = set()for code, meta in code_index.items():name = meta["name"]aliases = meta["aliases"]# 直接包含名称/别名或编码if code in query or name in query:direct_hits.add(code)else:# 任何别名命中for al in aliases.split(";"):if al and al in query:direct_hits.add(code)break# 混合检索候选docs = hybrid_retrieve(query, vectorstore, bm25, top_k_vec=40, top_k_bm25=40)# 如果规则命中,把这些 code 提到候选前列if direct_hits:prioritized = []seen = set()# 先加入直匹配for d in docs:c = d.metadata.get("code")if c in direct_hits and c not in seen:prioritized.append(d)seen.add(c)# 再补齐其他for d in docs:c = d.metadata.get("code")if c not in seen:prioritized.append(d)seen.add(c)docs = prioritized# 准备候选列表供 LLM 选择options, lookup = options_from_docs(docs, limit=top_candidates)allowed = set(lookup.keys())# 构造并调用 LLMprompt = SELECT_PROMPT.format(options=options, query=query)raw = llm(prompt)if isinstance(raw, dict) and "text" in raw:raw_text = raw["text"]else:# HF pipeline 一般直接返回strraw_text = str(raw)# 解析 LLM 输出picked = parse_selected_codes(raw_text, allowed_codes=allowed)# 合并直匹配与 LLM 选择(直匹配优先)final_codes = []seen = set()for c in list(direct_hits) + picked:if c not in seen:seen.add(c)final_codes.append(c)# 组装结果results = []evidences = {d.metadata.get("code"): d.page_content for d in docs}for c in final_codes:meta = code_index.get(c, {"code": c, "name": ""})results.append({"code": meta["code"],"name": meta["name"],"evidence": evidences.get(c, "")[:400] # 截取一小段证据文本})return {"query": query,"candidates_shown": options, # 便于调试查看"selected": results,"raw_model_output": raw_text}# ========== 示例入口 ==========
if __name__ == "__main__":excel_path = "ICD10.xlsx"logger.info("构建 ICD-10 知识库...")vectorstore, records, code_index = build_knowledge_base(excel_path=excel_path,embed_model="BAAI/bge-small-zh-v1.5", # 若下载不便,可替换为 "all-MiniLM-L6-v2"persist_dir="./icd10_chroma_db")logger.info("构建 BM25 检索器...")bm25 = build_bm25_retriever(records)logger.info("加载 DeepSeek 模型(低温度)...")llm = load_deepseek_model()user_text = ''' "患者女性,56岁,因「多饮多食伴消瘦1年,手足麻木2月」入院。""既往诊断 2 型糖尿病(未提及并发症),高血压 3 级(原发性,高危组),""否认其他疾病史。查体:空腹血糖 12.3mmol/L,下肢感觉减退..."'''logger.info("执行 编码匹配...")result = match_icd_codes(user_text=user_text,vectorstore=vectorstore,bm25=bm25,code_index=code_index,llm=llm,top_candidates=20)print("\n===== 结构化结果 =====")print(json.dumps(result["selected"], ensure_ascii=False, indent=2))print("\n===== 候选列表(诊断选择面板) =====")print(result["candidates_shown"])print("\n===== 原始模型输出(便于调试) =====")print(result["raw_model_output"])print(result["query"])结果如下:
2025-10-30 16:12:13,013 | INFO | 构建 ICD-10 知识库...
N:\数据分析\数据分析\deepseek_知识库改_gpt.py:107: LangChainDeprecationWarning: The class `HuggingFaceEmbeddings` was deprecated in LangChain 0.2.2 and will be removed in 1.0. An updated version of the class exists in the :class:`~langchain-huggingface package and should be used instead. To use it run `pip install -U :class:`~langchain-huggingface` and import as `from :class:`~langchain_huggingface import HuggingFaceEmbeddings``.embeddings = HuggingFaceEmbeddings(
2025-10-30 16:12:20,335 | INFO | Load pretrained SentenceTransformer: BAAI/bge-small-zh-v1.5
N:\数据分析\数据分析\deepseek_知识库改_gpt.py:121: LangChainDeprecationWarning: Since Chroma 0.4.x the manual persistence method is no longer supported as docs are automatically persisted.vectorstore.persist()
2025-10-30 16:13:45,908 | INFO | ✅ 向量库完成:./icd10_chroma_db,条目数=35862
2025-10-30 16:13:45,914 | INFO | 构建 BM25 检索器...
2025-10-30 16:13:47,330 | INFO | 加载 DeepSeek 模型(低温度)...
2025-10-30 16:13:52,449 | INFO | We will use 90% of the memory on device 0 for storing the model, and 10% for the buffer to avoid OOM. You can set `max_memory` in to a higher value to use more memory (at your own risk).
2025-10-30 16:14:07,573 | WARNING | Some parameters are on the meta device because they were offloaded to the cpu.
Device set to use cuda:0
N:\数据分析\数据分析\deepseek_知识库改_gpt.py:180: LangChainDeprecationWarning: The class `HuggingFacePipeline` was deprecated in LangChain 0.0.37 and will be removed in 1.0. An updated
version of the class exists in the :class:`~langchain-huggingface package and should be used instead. To use it run `pip install -U :class:`~langchain-huggingface` and import as `from :class:`~langchain_huggingface import HuggingFacePipeline``.return HuggingFacePipeline(pipeline=gen)
2025-10-30 16:14:07,578 | INFO | 执行 编码匹配...
N:\数据分析\数据分析\deepseek_知识库改_gpt.py:157: LangChainDeprecationWarning: The method `BaseRetriever.get_relevant_documents` was deprecated in langchain-core 0.1.46 and will be removed in 1.0. Use :meth:`~invoke` instead.bm25_docs = bm25.get_relevant_documents(query)
N:\数据分析\数据分析\deepseek_知识库改_gpt.py:299: LangChainDeprecationWarning: The method `BaseLLM.__call__` was deprecated in langchain-core 0.1.7 and will be removed in 1.0. Use :meth:`~invoke` instead.raw = llm(prompt)===== 结构化结果 =====
[{"code": "M12.801","name": "短暂性关节炎(病)","evidence": ""},{"code": "Q97.100","name": "女性,伴有多于三个X染色体的","evidence": ""},{"code": "C92.000x017","name": "急性髓细胞白血病,1/ETO型","evidence": ""},{"code": "L29.800","name": "瘙痒(症),其他的","evidence": ""},{"code": "L29.200","name": "外阴瘙痒(症)","evidence": ""},{"code": "Y17.x00","name": "气体和蒸气的中毒及暴露于该类物质,意图不确定的,其他","evidence": ""},{"code": "S92.203","name": "楔状骨骨折(足)","evidence": ""},{"code": "I10.x00x027","name": "高血压病2级(高危)","evidence": ""},{"code": "M11.801","name": "焦磷酸盐结晶性关节炎(病)","evidence": ""},{"code": "Y43.300","name": "抗肿瘤性药的有害效应,其他","evidence": ""},{"code": "I10.x00x031","name": "高血压病3级(高危)","evidence": ""},{"code": "R63.100x002","name": "多饮","evidence": ""},{"code": "R81.x00","name": "糖尿","evidence": ""},{"code": "M11.802","name": "磷酸二钙结晶性关节炎(病)","evidence": ""},{"code": "C97.x00","name": "独立(原发)多个部位的恶性肿瘤","evidence": ""},{"code": "I10.x00x002","name": "高血压","evidence": ""},{"code": "I10.x00x023","name": "高血压病1级(高危)","evidence": ""},{"code": "L29.000","name": "肛门瘙痒(症)","evidence": ""},{"code": "L29.100","name": "阴囊瘙痒(症)","evidence": ""},{"code": "J47.x00","name": "支气管扩张(症)","evidence": ""},{"code": "I10.x00","name": "特发性(原发性)高血压","evidence": ""},{"code": "M36.0*","name": "肿瘤病引起的皮(多)肌炎","evidence": ""},{"code": "R64.x00x002","name": "消瘦","evidence": ""},{"code": "R48.000","name": "诵读困难和失读(症)","evidence": ""},{"code": "L29.300","name": "肛门生殖器瘙痒(症)","evidence": ""},{"code": "L29.900","name": "瘙痒(症)","evidence": ""},{"code": "F64.000x002","name": "易性症,女","evidence": ""},{"code": "E14.900x001","name": "糖尿病","evidence": ""},{"code": "Q97.300","name": "女性,染色体伴有46XY核型","evidence": ""}
]===== 候选列表(诊断选择面板) =====
1) E11.406+G99.0* 2型糖尿病性胃轻瘫
2) A00.000 霍乱,由于O1群霍乱弧菌,霍乱生物型所致
3) E11.700x023 2型糖尿病性肥胖症性高血压
4) Y98.x00 与生活方式有关的情况
5) E11.400x350+G99.0* 2型糖尿病性食管功能障碍
6) Y97.x00 与环境污染有关的情况
7) E10.400x370+G99.0* 1型糖尿病性胃轻瘫
8) Y96.x00 与工作有关的情况
9) E11.503 2型糖尿病足病
10) Y95.x00 医源性情况
11) E11.600x012+M14.2* 2型糖尿病性手关节综合征
12) Y91.900 酒精影响
13) E11.500x050 2型糖尿病足
14) Y91.300 极严重的酒精中毒
15) E10.700x023 1型糖尿病性肥胖症性高血压
16) Y91.200 严重酒精中毒
17) E10.503 1型糖尿病性足病
18) Y91.100 中度酒精中毒
19) E14.400x370+G99.0* 糖尿病性胃轻瘫
20) Y91.000 轻度酒精中毒===== 原始模型输出(便于调试) =====
你是医学编码助手。你只能从候选列表中选择与“用户文本”明确匹配的诊断。禁止编造、禁止扩展、禁止输出候选列表之外的任何内容。
如果没有任何匹配,请输出空数组。候选列表(每行一个,格式:编号) 代码 名称):
1) E11.406+G99.0* 2型糖尿病性胃轻瘫
2) A00.000 霍乱,由于O1群霍乱弧菌,霍乱生物型所致
3) E11.700x023 2型糖尿病性肥胖症性高血压
4) Y98.x00 与生活方式有关的情况
5) E11.400x350+G99.0* 2型糖尿病性食管功能障碍
6) Y97.x00 与环境污染有关的情况
7) E10.400x370+G99.0* 1型糖尿病性胃轻瘫
8) Y96.x00 与工作有关的情况
9) E11.503 2型糖尿病足病
10) Y95.x00 医源性情况
11) E11.600x012+M14.2* 2型糖尿病性手关节综合征
12) Y91.900 酒精影响
13) E11.500x050 2型糖尿病足
14) Y91.300 极严重的酒精中毒
15) E10.700x023 1型糖尿病性肥胖症性高血压
16) Y91.200 严重酒精中毒
17) E10.503 1型糖尿病性足病
18) Y91.100 中度酒精中毒
19) E14.400x370+G99.0* 糖尿病性胃轻瘫
20) Y91.000 轻度酒精中毒用户文本:
"患者女性,56岁,因「多饮多食伴消瘦1年,手足麻木2月」入院。" "既往诊断 2 型糖尿病(未提及并发症),高血压 3 级(原发性,高危组)," "否认其他疾病史。查体:空腹血糖 12.3mmol/L,下肢感觉减退..." 严格输出 JSON,且仅包含一个键:
{"selected_codes": ["代码1","代码2", ...]}注意:
- 只能使用候选列表中的代码。
- 如果不确定或没有匹配,请输出 {"selected_codes": []}。
- 不要输出解释,不要输出多余文本。
- 请确保JSON格式正确,没有语法错误。
</think>```json
{"selected_codes": ["E11.406+G99.0*", "Y91.000"]}
结果分析:
按照之前豆包给的编码应该是这个:E11.9 → 2 型糖尿病 无并发症 I10.9 → 原发性高血压 3 级(高危组);但是上面结果是:E11.406+G99.0* 2型糖尿病性胃轻瘫,还输出了最后一个Y91.000 轻度酒精中毒;酒精中毒有点扯蛋了,2型糖尿病性胃轻瘫这个不清楚,感觉沾点边吧
小结
一文彻底搞懂大模型 - RAG(检索、增强、生成)
基本实现了RAG功能,当然,结果有待优化,后续更新吧!!
