【生物大模型文章精读实践七】HyenaDNA与Transformer的简单比较实践
接上期说要测试一下hyena和transformer的性能比较:
先看一下结果,目前没有看出差距,hyena代码参考的是作者的colab:https://colab.research.google.com/drive/1wyVEQd4R3HYLTUOXEEQmp_I8aNC_aLhL?usp=sharing
HyenaDNA_training_&inference_example(Public).ipynb 同时这部分代码没有测试hyena的强项,超长序列,还有待进一步检测:
Model Final val_loss Final val_ppl Best val_loss Last train_loss
------------------------------------------------------------------------
Hyena-S 3.7169 41.14 3.7137 3.7299
Hyena-M 3.7076 40.76 3.7103 3.7292
Transformer-S 3.7169 41.14 3.7091 3.7447
Transformer-M 3.7161 41.10 3.7175 3.7342--- Hyena-S ---
Final val_loss: 3.7169
Final val_ppl: 41.14
Best val_loss: 3.7137
Last train_loss: 3.7299
--- Hyena-M ---
Final val_loss: 3.7076
Final val_ppl: 40.76
Best val_loss: 3.7103
Last train_loss: 3.7292
--- Transformer-S ---
Final val_loss: 3.7169
Final val_ppl: 41.14
Best val_loss: 3.7091
Last train_loss: 3.7447
--- Transformer-M ---
Final val_loss: 3.7161
Final val_ppl: 41.10
Best val_loss: 3.7175
Last train_loss: 3.7342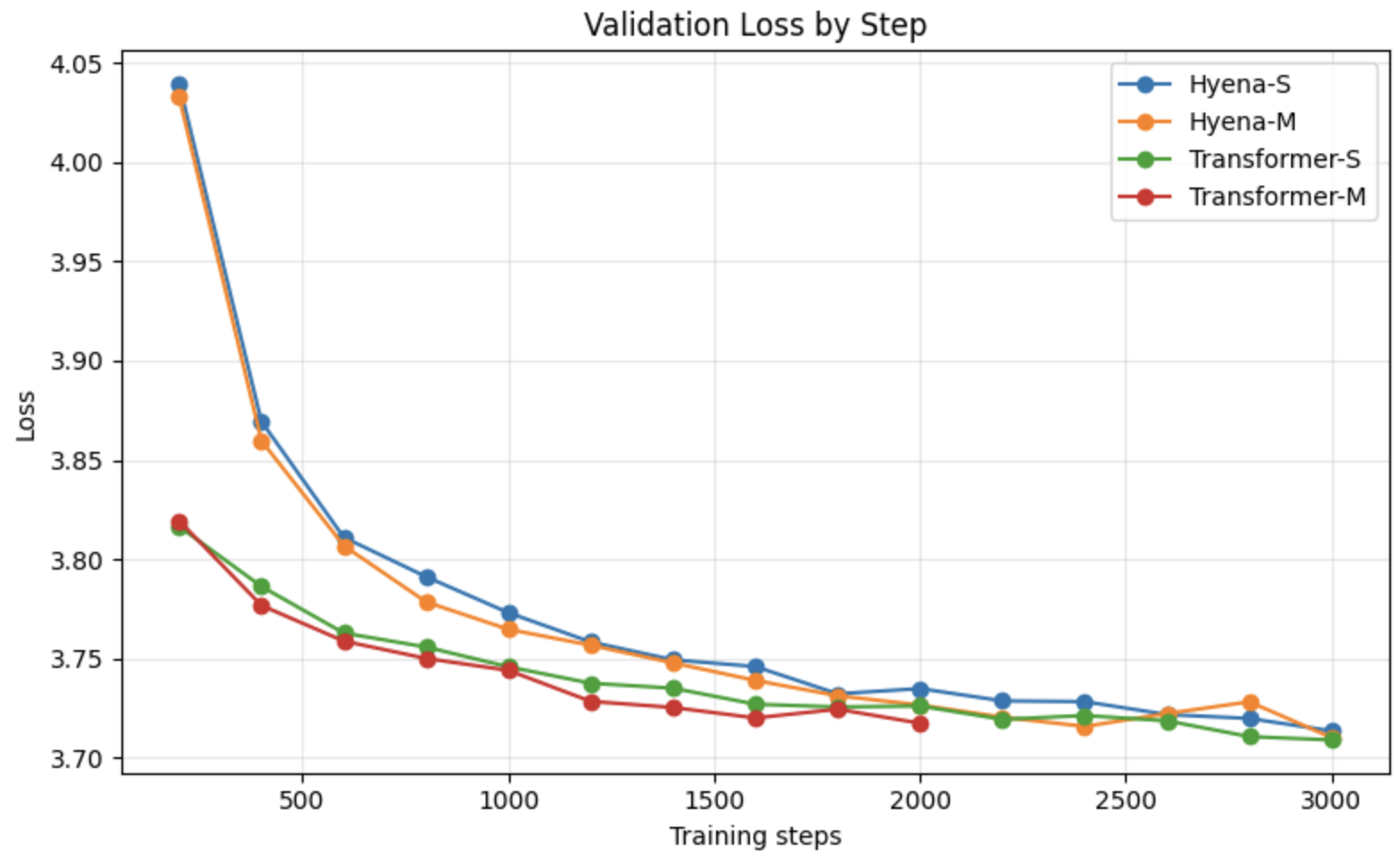
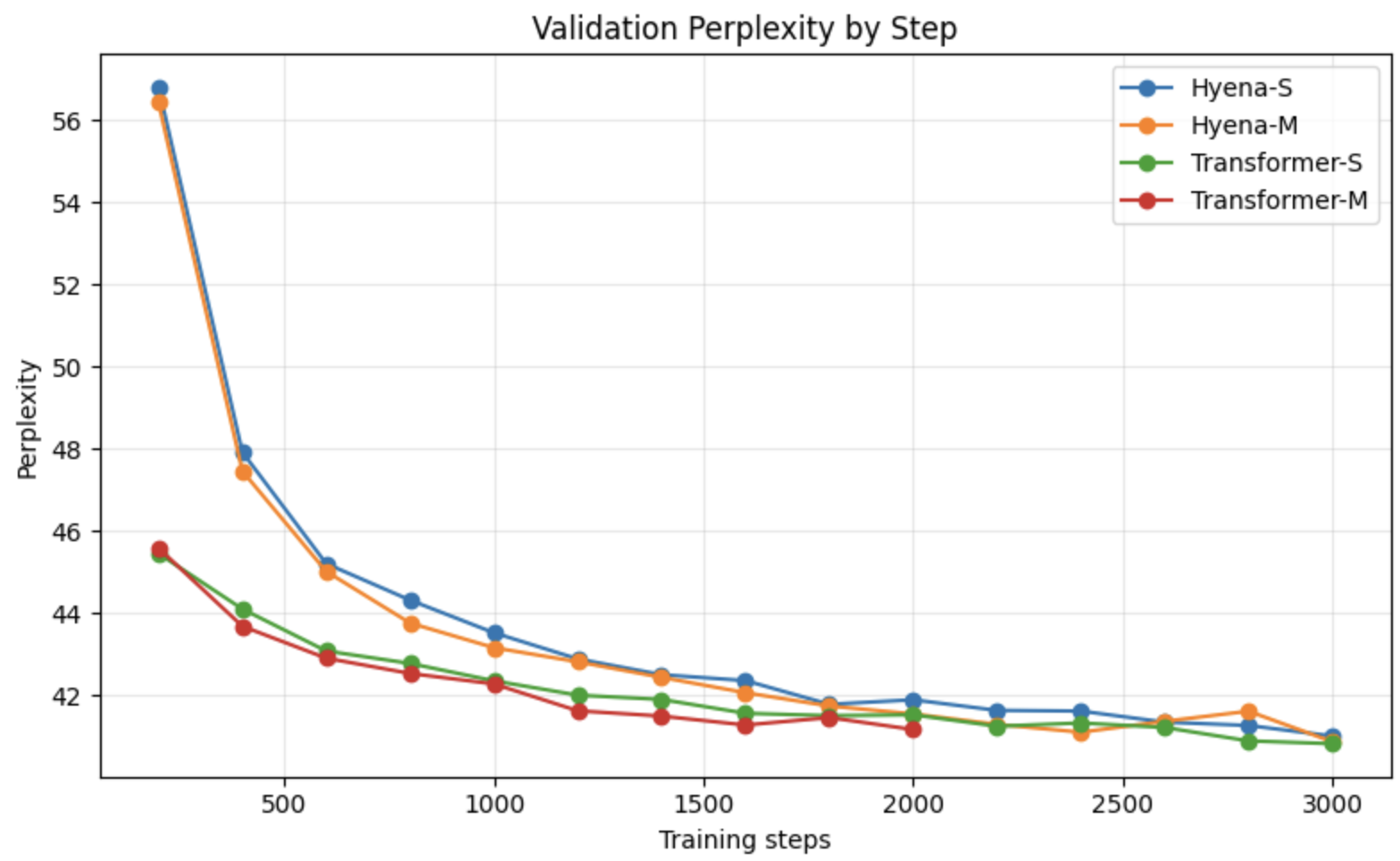
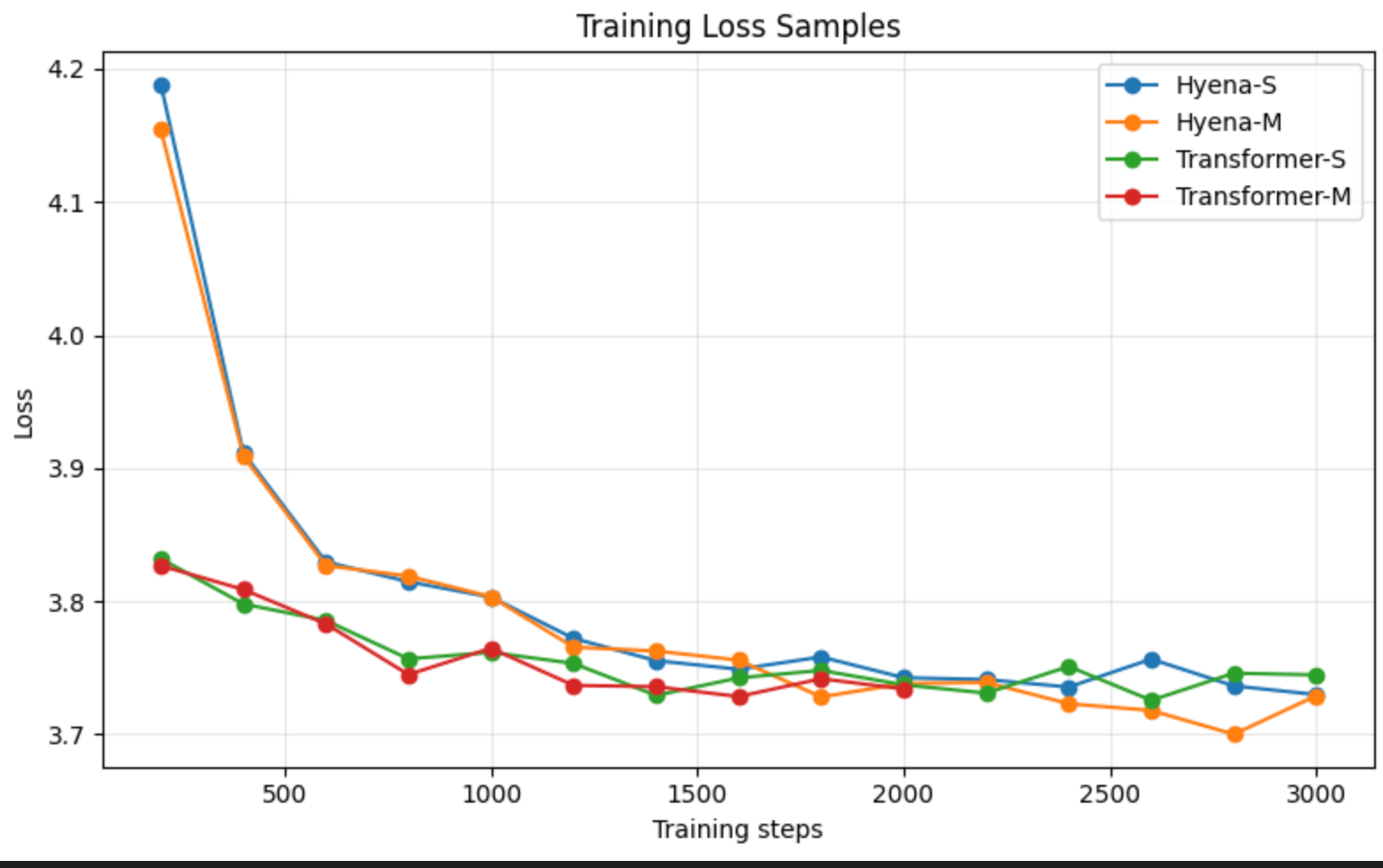
可以看出,虽然我的数据集很小,只有两个基因组,但是Transformer似乎比Hyena的loss下降快一些,但是最后两者2000step的下降loss差不多。
下面是架构和训练代码,供大家比较模型的时候参考 这是一下四个模型的参数
experiment_variants = {"Hyena-S": {"model_type": "hyena","d_model": 256,"n_layer": 4,"learning_rate": 2e-4,"grad_clip": 0.5,"total_steps": 3000,"mixed_precision": False,},"Hyena-M": {"model_type": "hyena","d_model": 384,"n_layer": 6,"order": 3,"filter_order": 96,"learning_rate": 1.5e-4,"grad_clip": 0.4,"total_steps": 3000,"mixed_precision": False,},"Transformer-S": {"model_type": "transformer","d_model": 256,"n_layer": 4,"n_head": 8,"learning_rate": 3e-4,"total_steps": 3000,},"Transformer-M": {"model_type": "transformer","d_model": 384,"n_layer": 6,"n_head": 8,"learning_rate": 2.5e-4,"total_steps": 2000,},}results: Dict[str, Dict[str, object]] = {}
for label, overrides in experiment_variants.items():print(f"===== Running {label} model =====")cfg = copy.deepcopy(shared_config)cfg.update(overrides)run_result = train_single_model(cfg, data_splits, device)results[label] = run_resultshared_config = {"data_dir": "./DNA","genome_files": ["Ruminococcus_albus.fna","Ruminococcus_flavefaciens.fna",],"kmer_size": 3,"train_split": 0.9,"min_orf_length": 90,"context_length": 512,"train_batch_size": 32,"eval_batch_size": 32,"total_steps": 2000,"learning_rate": 6e-4,"weight_decay": 0.1,"grad_clip": 1.0,"seed": 2222,"d_model": 256,"n_layer": 6,"n_head": 8,"order": 3,"filter_order": 64,"dropout": 0.1,"mixed_precision": True,"log_interval": 50,"eval_interval": 200,"eval_steps": 20,"save_checkpoint": False,"checkpoint_dir": "./checkpoints","use_coding_sequences": True,"prefer_mps": False,
}set_seed(shared_config["seed"])
device = select_device(shared_config.get("prefer_mps", False))
print(f"Using device: {device}")
if device.type == "mps":print("MPS detected. Hyena FFT uses CPU fallback because torch.fft is not implemented on mps.")
import os
os.environ.setdefault("PYTORCH_ENABLE_MPS_FALLBACK", "1")import copy
import math
import random
import re
from pathlib import Path
from typing import Dict, List, Optional, Sequence, Tupleimport torch
import torch.nn as nn
import torch.nn.functional as F
from torch.cuda.amp import GradScaler, autocast
from tqdm.auto import tqdm
import matplotlib.pyplot as plt
from einops import rearrangedef set_seed(seed: int) -> None:random.seed(seed)torch.manual_seed(seed)if torch.cuda.is_available():torch.cuda.manual_seed_all(seed)def count_parameters(module: nn.Module) -> int:return sum(p.numel() for p in module.parameters() if p.requires_grad)def select_device(prefer_mps: bool = False) -> torch.device:if torch.cuda.is_available():return torch.device("cuda")use_mps = getattr(torch.backends, "mps", None) and torch.backends.mps.is_available()if use_mps and prefer_mps:return torch.device("mps")return torch.device("cpu")# Hyena building blocks (from the public tutorial, expressed as modules)import torchdef fftconv(u, k, D):"""Apply convolution via the Fourier domain (MPS-safe)."""seqlen = u.shape[-1]fft_size = 2 * seqlenorig_device = u.deviceorig_dtype = u.dtypecompute_device = torch.device("cpu") if orig_device.type == "mps" else orig_deviceu_work = u.to(compute_device, dtype=k.dtype)k_work = k.to(compute_device)D_work = D.to(compute_device)k_f = torch.fft.rfft(k_work, n=fft_size) / fft_sizeu_f = torch.fft.rfft(u_work, n=fft_size)if len(u.shape) > 3:k_f = k_f.unsqueeze(1)y = torch.fft.irfft(u_f * k_f, n=fft_size, norm="forward")[..., :seqlen]out = y + u_work * D_work.unsqueeze(-1)return out.to(orig_device, dtype=orig_dtype)@torch.jit.script
def mul_sum(q, y):return (q * y).sum(dim=1)class OptimModule(nn.Module):"""Module helper to register tensors with custom optimizer hyperparameters."""def register(self, name, tensor, lr=None, wd=0.0):if lr == 0.0:self.register_buffer(name, tensor)else:self.register_parameter(name, nn.Parameter(tensor))optim = {}if lr is not None:optim["lr"] = lrif wd is not None:optim["weight_decay"] = wdsetattr(getattr(self, name), "_optim", optim)class Sin(nn.Module):"""Sinusoidal activation for the Hyena filter MLP."""def __init__(self, dim: int, w: float = 10.0, train_freq: bool = True):super().__init__()if train_freq:self.freq = nn.Parameter(w * torch.ones(1, dim))else:self.freq = w * torch.ones(1, dim)def forward(self, x: torch.Tensor) -> torch.Tensor:return torch.sin(self.freq * x)class PositionalEmbedding(OptimModule):def __init__(self, emb_dim: int, seq_len: int, lr_pos_emb: float = 1e-5, **kwargs):super().__init__()self.seq_len = seq_lent = torch.linspace(0, 1, self.seq_len)[None, :, None]if emb_dim > 1:bands = (emb_dim - 1) // 2t_rescaled = torch.linspace(0, seq_len - 1, seq_len)[None, :, None]w = 2 * math.pi * t_rescaled / seq_lenf = torch.linspace(1e-4, bands - 1, bands)[None, None]z = torch.exp(-1j * f * w)z = torch.cat([t, z.real, z.imag], dim=-1)self.register("z", z, lr=lr_pos_emb)self.register("t", t, lr=0.0)def forward(self, L: int):return self.z[:, :L], self.t[:, :L]class ExponentialModulation(OptimModule):def __init__(self,d_model: int,fast_decay_pct: float = 0.3,slow_decay_pct: float = 1.5,target: float = 1e-2,modulation_lr: float = 0.0,modulate: bool = True,shift: float = 0.05,**kwargs,):super().__init__()self.modulate = modulateself.shift = shiftmax_decay = math.log(target) / fast_decay_pctmin_decay = math.log(target) / slow_decay_pctdeltas = torch.linspace(min_decay, max_decay, d_model)[None, None]self.register("deltas", deltas, lr=modulation_lr)def forward(self, t: torch.Tensor, x: torch.Tensor) -> torch.Tensor:if self.modulate:decay = torch.exp(-t * self.deltas.abs())x = x * (decay + self.shift)return xclass HyenaFilter(OptimModule):def __init__(self,d_model: int,emb_dim: int = 3,order: int = 16,fused_fft_conv: bool = False,seq_len: int = 1024,lr: float = 1e-3,lr_pos_emb: float = 1e-5,dropout: float = 0.0,w: float = 1.0,wd: float = 0.0,bias: bool = True,num_inner_mlps: int = 2,normalized: bool = False,**kwargs,):super().__init__()self.d_model = d_modelself.use_bias = biasself.fused_fft_conv = fused_fft_convself.bias = nn.Parameter(torch.randn(self.d_model))self.dropout = nn.Dropout(dropout)act = Sin(dim=order, w=w)self.emb_dim = emb_dimassert emb_dim % 2 != 0 and emb_dim >= 3self.seq_len = seq_lenself.pos_emb = PositionalEmbedding(emb_dim, seq_len, lr_pos_emb)implicit_filter = [nn.Linear(emb_dim, order), act]for _ in range(num_inner_mlps):implicit_filter.extend([nn.Linear(order, order), act])implicit_filter.append(nn.Linear(order, d_model, bias=False))self.implicit_filter = nn.Sequential(*implicit_filter)self.modulation = ExponentialModulation(d_model, **kwargs)self.normalized = normalizedfor child in self.implicit_filter.children():for name, _ in child.state_dict().items():optim = {"weight_decay": wd, "lr": lr}setattr(getattr(child, name), "_optim", optim)def filter(self, L: int, *args, **kwargs):z, t = self.pos_emb(L)h = self.implicit_filter(z)h = self.modulation(t, h)return hdef forward(self, x: torch.Tensor, L: int, k: Optional[torch.Tensor] = None, bias: Optional[torch.Tensor] = None,*args, **kwargs) -> torch.Tensor:if k is None:k = self.filter(L)k = k[0] if isinstance(k, tuple) else ky = fftconv(x, k, bias)return yclass HyenaOperator(nn.Module):def __init__(self,d_model: int,l_max: int,order: int = 2,filter_order: int = 64,dropout: float = 0.0,filter_dropout: float = 0.0,**filter_args,):super().__init__()self.d_model = d_modelself.l_max = l_maxself.order = orderinner_width = d_model * (order + 1)self.dropout = nn.Dropout(dropout)self.in_proj = nn.Linear(d_model, inner_width)self.out_proj = nn.Linear(d_model, d_model)self.short_filter = nn.Conv1d(inner_width, inner_width, 3, padding=2, groups=inner_width)self.filter_fn = HyenaFilter(d_model * (order - 1),order=filter_order,seq_len=l_max,channels=1,dropout=filter_dropout,**filter_args,)def forward(self, u: torch.Tensor, *args, **kwargs) -> torch.Tensor:l = u.size(-2)l_filter = min(l, self.l_max)u = self.in_proj(u)u = rearrange(u, 'b l d -> b d l')uc = self.short_filter(u)[..., :l_filter]*x, v = uc.split(self.d_model, dim=1)k = self.filter_fn.filter(l_filter)[0]k = rearrange(k, 'l (o d) -> o d l', o=self.order - 1)bias = rearrange(self.filter_fn.bias, '(o d) -> o d', o=self.order - 1)for o, x_i in enumerate(reversed(x[1:])):v = self.dropout(v * x_i)v = self.filter_fn(v, l_filter, k=k[o], bias=bias[o])y = rearrange(v * x[0], 'b d l -> b l d')return self.out_proj(y)class HyenaBlock(nn.Module):def __init__(self, d_model: int, l_max: int, order: int, filter_order: int, dropout: float):super().__init__()self.norm = nn.LayerNorm(d_model)self.hyena = HyenaOperator(d_model=d_model,l_max=l_max,order=order,filter_order=filter_order,dropout=dropout,)self.mlp = nn.Sequential(nn.LayerNorm(d_model),nn.Linear(d_model, 4 * d_model),nn.GELU(),nn.Linear(4 * d_model, d_model),nn.Dropout(dropout),)def forward(self, x: torch.Tensor) -> torch.Tensor:residual = xx = self.norm(x)x = self.hyena(x)x = x + residualx = x + self.mlp(x)return xclass HyenaBackbone(nn.Module):def __init__(self,vocab_size: int,d_model: int,n_layer: int,l_max: int,order: int,filter_order: int,dropout: float,):super().__init__()self.embedding = nn.Embedding(vocab_size, d_model)self.layers = nn.ModuleList([HyenaBlock(d_model, l_max, order, filter_order, dropout) for _ in range(n_layer)])self.norm = nn.LayerNorm(d_model)def forward(self, x: torch.Tensor) -> torch.Tensor:x = self.embedding(x)for layer in self.layers:x = layer(x)return self.norm(x)class HyenaLanguageModel(nn.Module):def __init__(self, backbone: HyenaBackbone, tie_weights: bool = True):super().__init__()self.backbone = backboneself.lm_head = nn.Linear(backbone.embedding.embedding_dim,backbone.embedding.num_embeddings,bias=False,)if tie_weights:self.lm_head.weight = self.backbone.embedding.weightdef forward(self, x: torch.Tensor) -> torch.Tensor:hidden = self.backbone(x)return self.lm_head(hidden)# Transformer baseline modules for comparison
class CausalSelfAttention(nn.Module):def __init__(self, d_model: int, n_head: int, dropout: float, context_length: int):super().__init__()if d_model % n_head != 0:raise ValueError(f"d_model={d_model} must be divisible by n_head={n_head}")self.n_head = n_headself.head_dim = d_model // n_headself.qkv = nn.Linear(d_model, 3 * d_model)self.out_proj = nn.Linear(d_model, d_model)self.attn_drop = nn.Dropout(dropout)self.resid_drop = nn.Dropout(dropout)mask = torch.tril(torch.ones(context_length, context_length)).view(1, 1, context_length, context_length)self.register_buffer('mask', mask)def forward(self, x: torch.Tensor) -> torch.Tensor:B, T, C = x.size()qkv = self.qkv(x)q, k, v = qkv.chunk(3, dim=-1)q = q.view(B, T, self.n_head, self.head_dim).transpose(1, 2)k = k.view(B, T, self.n_head, self.head_dim).transpose(1, 2)v = v.view(B, T, self.n_head, self.head_dim).transpose(1, 2)att = (q @ k.transpose(-2, -1)) / math.sqrt(self.head_dim)mask = self.mask[:, :, :T, :T]att = att.masked_fill(mask == 0, float('-inf'))att = torch.softmax(att, dim=-1)att = self.attn_drop(att)y = att @ vy = y.transpose(1, 2).contiguous().view(B, T, C)y = self.resid_drop(self.out_proj(y))return yclass TransformerBlock(nn.Module):def __init__(self, d_model: int, n_head: int, dropout: float, context_length: int):super().__init__()self.ln1 = nn.LayerNorm(d_model)self.ln2 = nn.LayerNorm(d_model)self.attn = CausalSelfAttention(d_model, n_head, dropout, context_length)self.mlp = nn.Sequential(nn.Linear(d_model, 4 * d_model),nn.GELU(),nn.Linear(4 * d_model, d_model),nn.Dropout(dropout),)def forward(self, x: torch.Tensor) -> torch.Tensor:x = x + self.attn(self.ln1(x))x = x + self.mlp(self.ln2(x))return xclass TransformerLanguageModel(nn.Module):def __init__(self, vocab_size: int, d_model: int, n_layer: int, n_head: int, context_length: int, dropout: float):super().__init__()self.block_size = context_lengthself.token_emb = nn.Embedding(vocab_size, d_model)self.pos_emb = nn.Parameter(torch.zeros(1, context_length, d_model))self.drop = nn.Dropout(dropout)self.blocks = nn.ModuleList([TransformerBlock(d_model, n_head, dropout, context_length) for _ in range(n_layer)])self.ln_f = nn.LayerNorm(d_model)self.head = nn.Linear(d_model, vocab_size, bias=False)self.apply(self._init_weights)def _init_weights(self, module):if isinstance(module, nn.Linear):nn.init.normal_(module.weight, mean=0.0, std=0.02)if module.bias is not None:nn.init.zeros_(module.bias)elif isinstance(module, nn.Embedding):nn.init.normal_(module.weight, mean=0.0, std=0.02)def forward(self, idx: torch.Tensor) -> torch.Tensor:B, T = idx.size()if T > self.block_size:raise ValueError(f"Sequence length {T} exceeds block_size {self.block_size}")tok = self.token_emb(idx)pos = self.pos_emb[:, :T, :]x = self.drop(tok + pos)for block in self.blocks:x = block(x)x = self.ln_f(x)return self.head(x)# DNA preprocessing helpers for autoregressive modeling
SEP_TOKEN = "<SEP>"
complement_map = str.maketrans({"A": "T", "T": "A", "C": "G", "G": "C", "N": "N"})
START_CODONS = {"ATG", "GTG", "TTG"}
STOP_CODONS = {"TAA", "TAG", "TGA"}def read_fasta_sequences(path: Path) -> Dict[str, str]:sequences: Dict[str, str] = {}header: Optional[str] = Nonechunks: List[str] = []with open(path, "r", encoding="utf-8") as handle:for line in handle:line = line.strip()if not line:continueif line.startswith(">"):if header is not None:sequences[header] = "".join(chunks).upper()header = line[1:].split()[0]chunks = []else:chunks.append(line)if header is not None:sequences[header] = "".join(chunks).upper()return sequencesdef clean_sequence(seq: str) -> str:cleaned = "".join(ch for ch in seq.upper() if ch in "ACGTN")return re.sub(r"N{6,}", "NNNNN", cleaned)def reverse_complement(seq: str) -> str:return seq.translate(complement_map)[::-1]def find_orfs(seq: str, min_len: int) -> List[str]:orfs: List[str] = []length = len(seq)for frame in range(3):i = framewhile i + 3 <= length:codon = seq[i : i + 3]if codon in START_CODONS:j = i + 3while j + 3 <= length:stop = seq[j : j + 3]if stop in STOP_CODONS:orf_len = j + 3 - iif orf_len >= min_len:orfs.append(seq[i : j + 3])breakj += 3i = jelse:i += 3return orfsdef extract_orfs_from_genomes(genome_paths: Sequence[Path], min_len: int) -> Tuple[Dict[str, Dict[str, str]], List[str]]:genome_sequences: Dict[str, Dict[str, str]] = {}cds_sequences: List[str] = []for path in genome_paths:sequences = read_fasta_sequences(path)genome_sequences[path.name] = sequencesfor seqname, seq in sequences.items():cleaned = clean_sequence(seq)cds_sequences.extend(find_orfs(cleaned, min_len=min_len))rc = reverse_complement(cleaned)cds_sequences.extend(find_orfs(rc, min_len=min_len))return genome_sequences, cds_sequencesdef collect_kmers(text: str, k: int) -> set:kmers: set = set()if len(text) < k:return kmersfor i in range(0, len(text) - k + 1):chunk = text[i : i + k]if "<" in chunk or ">" in chunk:continuekmers.add(chunk)return kmersdef build_data_tensors(token_ids: Sequence[int], split_ratio: float) -> Dict[str, torch.Tensor]:encoded = torch.tensor(token_ids, dtype=torch.long)split_idx = int(split_ratio * encoded.numel())return {"train": encoded[:split_idx],"val": encoded[split_idx:],}def get_batch(data_dict: Dict[str, torch.Tensor], split: str, block_size: int, batch_size: int) -> Tuple[torch.Tensor, torch.Tensor]:data = data_dict[split]if data.size(0) <= block_size + 1:raise ValueError(f"Data too short for block_size={block_size}, length={data.size(0)}")max_start = data.size(0) - block_size - 1idx = torch.randint(0, max_start, (batch_size,))x = torch.stack([data[i : i + block_size] for i in idx])y = torch.stack([data[i + 1 : i + block_size + 1] for i in idx])return x, y@torch.no_grad()
def evaluate(model: nn.Module, data_dict: Dict[str, torch.Tensor], device: torch.device, *, block_size: int, batch_size: int, steps: int) -> Tuple[float, float]:model.eval()losses: List[float] = []for _ in range(steps):xb, yb = get_batch(data_dict, "val", block_size, batch_size)xb, yb = xb.to(device), yb.to(device)logits = model(xb)loss = F.cross_entropy(logits.view(-1, logits.size(-1)), yb.view(-1))losses.append(loss.item())avg_loss = sum(losses) / max(len(losses), 1)return avg_loss, math.exp(avg_loss)data_dir = Path(shared_config["data_dir"])
genome_paths = [data_dir / name for name in shared_config["genome_files"]]
print("Loading genomes:", genome_paths)genome_sequences, cds_sequences = extract_orfs_from_genomes(genome_paths, shared_config["min_orf_length"])
print(f"Loaded {len(genome_sequences)} genomes, ORFs extracted: {len(cds_sequences)}")if not cds_sequences:raise RuntimeError("No ORFs found. Adjust min_orf_length or check genome files.")coding_text = SEP_TOKEN.join(cds_sequences)
flat_sequences = [clean_sequence(seq)for genome in genome_sequences.values()for seq in genome.values()
]
full_genome_text = SEP_TOKEN.join(flat_sequences)print(f"Coding characters: {len(coding_text)}")
print(f"Genome characters: {len(full_genome_text)}")corpus_text = coding_text if shared_config["use_coding_sequences"] else full_genome_text
corpus_name = "coding ORFs" if shared_config["use_coding_sequences"] else "full genomes"
print(f"Training corpus: {corpus_name}")allowed_chars = {"A", "C", "G", "T", "N"}
dna_chars = {ch for ch in corpus_text if ch in allowed_chars}
single_tokens = sorted(dna_chars | {SEP_TOKEN})
kmer_tokens = sorted(collect_kmers(coding_text, shared_config["kmer_size"]) | collect_kmers(full_genome_text, shared_config["kmer_size"])
)vocab = single_tokens + [tok for tok in kmer_tokens if tok not in single_tokens]
stoi = {tok: idx for idx, tok in enumerate(vocab)}
itos = {idx: tok for tok, idx in stoi.items()}
kmer_token_set = set(kmer_tokens)print(f"Single tokens: {len(single_tokens)}")
print(f"K-mer tokens: {len(kmer_tokens)}")
print(f"Vocab size: {len(vocab)}")def encode(text: str) -> List[int]:ids: List[int] = []i = 0length = len(text)sep_len = len(SEP_TOKEN)while i < length:if text.startswith(SEP_TOKEN, i):ids.append(stoi[SEP_TOKEN])i += sep_lencontinueif i + shared_config['kmer_size'] <= length:chunk = text[i : i + shared_config['kmer_size']]if "<" not in chunk and chunk in kmer_token_set:ids.append(stoi[chunk])i += shared_config['kmer_size']continuetoken = text[i]if token not in stoi:token = "N"ids.append(stoi[token])i += 1return idsdef decode(token_ids: Sequence[int]) -> str:return "".join(itos[int(i)] for i in token_ids)def decode_to_text(token_ids) -> str:if isinstance(token_ids, torch.Tensor):token_ids = token_ids.tolist()if token_ids and isinstance(token_ids[0], list):token_ids = token_ids[0]return decode(token_ids)encoded_ids = encode(corpus_text)
data_splits = build_data_tensors(encoded_ids, shared_config["train_split"])print(f"Train tokens: {data_splits['train'].numel()}")
print(f"Val tokens: {data_splits['val'].numel()}")shared_config["vocab_size"] = len(vocab)def build_model(config: Dict[str, object], device: torch.device) -> nn.Module:model_type = config["model_type"].lower()if model_type == "hyena":backbone = HyenaBackbone(vocab_size=config["vocab_size"],d_model=config["d_model"],n_layer=config["n_layer"],l_max=config["context_length"],order=config["order"],filter_order=config["filter_order"],dropout=config["dropout"],)model = HyenaLanguageModel(backbone)elif model_type == "transformer":model = TransformerLanguageModel(vocab_size=config["vocab_size"],d_model=config["d_model"],n_layer=config["n_layer"],n_head=config["n_head"],context_length=config["context_length"],dropout=config["dropout"],)else:raise ValueError(f"Unsupported model_type {config['model_type']}")return model.to(device)def train_single_model(config: Dict[str, object], data_splits: Dict[str, torch.Tensor], device: torch.device) -> Dict[str, object]:config = copy.deepcopy(config)set_seed(config["seed"])model = build_model(config, device)print(f"Model parameters: {count_parameters(model):,}")optimizer = torch.optim.AdamW(model.parameters(), lr=config["learning_rate"], weight_decay=config["weight_decay"])scaler = GradScaler(enabled=config["mixed_precision"] and device.type == "cuda")history: List[Dict[str, float]] = []best_val_loss = float("inf")last_train_loss = Noneprogress_bar = tqdm(range(1, config["total_steps"] + 1),desc=f"{config['model_type'].title()} Training",total=config["total_steps"],)for step in progress_bar:model.train()xb, yb = get_batch(data_splits, "train", config["context_length"], config["train_batch_size"])xb, yb = xb.to(device), yb.to(device)optimizer.zero_grad(set_to_none=True)with autocast(enabled=config["mixed_precision"] and device.type == "cuda"):logits = model(xb)loss = F.cross_entropy(logits.view(-1, logits.size(-1)), yb.view(-1))loss_value = loss.item()last_train_loss = loss_valueif scaler.is_enabled():scaler.scale(loss).backward()if config["grad_clip"] is not None:scaler.unscale_(optimizer)torch.nn.utils.clip_grad_norm_(model.parameters(), config["grad_clip"])scaler.step(optimizer)scaler.update()else:loss.backward()if config["grad_clip"] is not None:torch.nn.utils.clip_grad_norm_(model.parameters(), config["grad_clip"])optimizer.step()if config["log_interval"] and step % config["log_interval"] == 0:progress_bar.set_postfix(train_loss=f"{loss_value:.4f}", refresh=False)if config["eval_interval"] and step % config["eval_interval"] == 0:val_loss, val_ppl = evaluate(model,data_splits,device,block_size=config["context_length"],batch_size=config["eval_batch_size"],steps=config["eval_steps"],)history.append({"step": step, "train_loss": loss_value, "val_loss": val_loss, "val_ppl": val_ppl})print(f"[Eval] step {step:>5d} | val_loss {val_loss:.4f} | val_ppl {val_ppl:.2f}")if val_loss < best_val_loss:best_val_loss = val_lossif config.get("save_checkpoint"):checkpoint_dir = Path(config.get("checkpoint_dir", "."))checkpoint_dir.mkdir(parents=True, exist_ok=True)checkpoint_path = checkpoint_dir / f"{config['model_type']}_comparison.pt"torch.save({"model": model.state_dict(), "config": config}, checkpoint_path)print(f"Checkpoint saved to {checkpoint_path}")final_val_loss, final_val_ppl = evaluate(model,data_splits,device,block_size=config["context_length"],batch_size=config["eval_batch_size"],steps=config["eval_steps"],)print(f"Final val_loss: {final_val_loss:.4f}, val_ppl: {final_val_ppl:.2f}")return {"model": model,"history": history,"best_val_loss": best_val_loss,"final_val_loss": final_val_loss,"final_val_ppl": final_val_ppl,"last_train_loss": last_train_loss,"config": config,}@torch.no_grad()
def sample_autoregressive(model: nn.Module,start_tokens: torch.Tensor,max_new_tokens: int,temperature: float = 1.0,top_k: Optional[int] = None,
) -> torch.Tensor:model.eval()generated = start_tokens.clone()for _ in range(max_new_tokens):idx_cond = generated[:, -start_tokens.size(1):]logits = model(idx_cond)logits = logits[:, -1, :] / max(temperature, 1e-6)if top_k is not None:values, indices = torch.topk(logits, top_k)mask = torch.full_like(logits, float('-inf'))mask.scatter_(1, indices, values)logits = maskprobs = torch.softmax(logits, dim=-1)next_token = torch.multinomial(probs, num_samples=1)generated = torch.cat((generated, next_token), dim=1)return generated# Visualization helpersdef aggregate_histories(results: Dict[str, Dict[str, object]]) -> Dict[str, Dict[str, object]]:aggregated: Dict[str, Dict[str, object]] = {}for label, payload in results.items():history = payload.get("history") or []aggregated[label] = {"steps": [entry.get("step") for entry in history if entry.get("step") is not None],"train_loss": [entry.get("train_loss") for entry in history if entry.get("train_loss") is not None],"val_loss": [entry.get("val_loss") for entry in history if entry.get("val_loss") is not None],"val_ppl": [entry.get("val_ppl") for entry in history if entry.get("val_ppl") is not None],"final": {"val_loss": payload.get("final_val_loss"),"val_ppl": payload.get("final_val_ppl"),"best_val_loss": payload.get("best_val_loss"),"last_train_loss": payload.get("last_train_loss"),},}return aggregateddef plot_metric_curves(aggregated: Dict[str, Dict[str, object]], metric: str, *, title: str, ylabel: str) -> None:plt.figure(figsize=(8, 5))plotted = Falsefor label, data in aggregated.items():steps = data.get("steps") or []values = data.get(metric) or []if steps and values and len(steps) == len(values):plt.plot(steps, values, marker="o", label=label)plotted = Trueif not plotted:plt.close()print(f"No data available to plot {metric} curves.")returnplt.title(title)plt.xlabel("Training steps")plt.ylabel(ylabel)plt.grid(True, alpha=0.3)plt.legend()plt.tight_layout()plt.show()def plot_final_metric_bar(results: Dict[str, Dict[str, object]], key: str, *, title: str, ylabel: str) -> None:labels = []values = []for label, payload in results.items():value = payload.get(f"final_{key}") if not key.startswith("final_") else payload.get(key)if value is None:value = payload.get(key)if value is None:continuelabels.append(label)values.append(value)if not values:print(f"No final values available for {key}.")returnplt.figure(figsize=(8, 5))bars = plt.bar(labels, values)plt.title(title)plt.ylabel(ylabel)plt.grid(axis="y", alpha=0.3)plt.xticks(rotation=30, ha="right")for bar, value in zip(bars, values):plt.text(bar.get_x() + bar.get_width() / 2, bar.get_height(), f"{value:.3f}", ha="center", va="bottom", fontsize=9)plt.tight_layout()plt.show()aggregated = aggregate_histories(results)plot_metric_curves(aggregated, 'val_loss', title='Validation Loss by Step', ylabel='Loss')
plot_metric_curves(aggregated, 'val_ppl', title='Validation Perplexity by Step', ylabel='Perplexity')
plot_metric_curves(aggregated, 'train_loss', title='Training Loss Samples', ylabel='Loss')plot_final_metric_bar(results, 'final_val_loss', title='Final Validation Loss Comparison', ylabel='Loss')
plot_final_metric_bar(results, 'final_val_ppl', title='Final Validation Perplexity Comparison', ylabel='Perplexity')