LightDock:高效蛋白质-DNA对接框架
GitHub - lightdock/lightdock: Protein-protein, protein-peptide and protein-DNA docking framework based on the GSO algorithm
lightdock - 大分子对接框架_PyPI中文网
蛋白质-DNA 对接 | LightDock --- Protein-DNA Docking | LightDock
先学会后应用~
1.下载安装相关包~
lightdock
pip3 install lightdock -i https://pypi.tuna.tsinghua.edu.cn/simplegit clone https://github.com/lightdock/lightdock.gitcd lightdock
pip install -e .
# LightDock
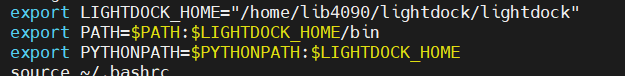
export LIGHTDOCK_HOME="/path/to/lightdock"
export PATH=$PATH:$LIGHTDOCK_HOME/bin
export PYTHONPATH=$PYTHONPATH:$LIGHTDOCK_HOME
reduce
git clone https://github.com/rlabduke/reduce.git使用cmake
mkdir -p ~/build/reduce
cd ~/build/reduce
cmake ~/src/reduce
make
sudo make installPDB-Tools
Python 包 GitHub - haddocking/pdb-tools: A dependency-free cross-platform swiss army knife for PDB files.
pip install pdb-tools# To download
git clone https://github.com/haddocking/pdb-tools
cd pdb-tools# To update
git pull origin master# To install
python setup.py install
2.linux run LightDock
以官方示例数据作为演示
- 1. Copying the data
-
mkdir data cd data curl -O https://raw.githubusercontent.com/lightdock/lightdock.github.io/master/tutorials/0.9.3/dna_docking/data/1AZP_A.pdb curl -O https://raw.githubusercontent.com/lightdock/lightdock.github.io/master/tutorials/0.9.3/dna_docking/data/1AZP_B.pdb - 2. Specifying residue restraints
-
cd lightdock/lightdock/lightdock/curl -O https://raw.githubusercontent.com/lightdock/lightdock.github.io/master/tutorials/0.9.3/dna_docking/data/restraints.list - 3. Protonation
- 3.1. Protein
-
reduce -Trim 1AZP_A.pdb > 1AZP_A_noh.pdb reduce -BUILD 1AZP_A_noh.pdb > 1AZP_A_h.pdb -
pdb_atom_renumber 1AZP_A_h.pdb > protein.pdb Renumbering a structure 重新编号结构pdb_reres -1 1AZP_A_h.pdb > protein.pdb
3.2. DNA-
reduce -Trim 1AZP_B.pdb > 1AZP_B_noh.pdb reduce -BUILD 1AZP_B_noh.pdb > 1AZP_B_h.pdb上传 reduce_to_amber.py 到 lightdock/lightdock/lightdock/
-
mv data/1AZP_B_h.pdb ../lightdock/cd lightdock/lightdock/lightdock/ python reduce_to_amber.py 1AZP_B_h.pdb dna.pdb
- 4. Setup
-
mv data/protein.pdb ../lightdocklightdock3_setup.py protein.pdb dna.pdb -anm -rst restraints.listlgd_setup.py ./lightdock/protein.pdb ./lightdock/dna.pdb -anm -rst ./lightdock/restraints.list 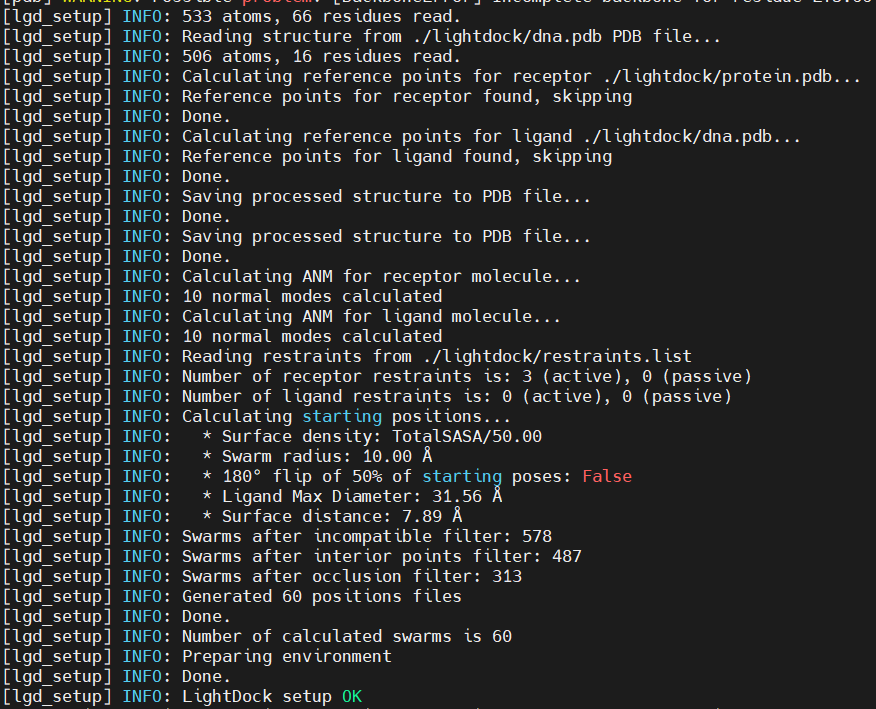
- 5. Simulation
-
lightdock3.py setup.json 100 -s dna -c 8lgd_run.py setup.json 100 -s dna -c 8 - 6. Clustering and Filtering
-
#PBS -N 1AZP-post #PBS -q medium #PBS -l nodes=1:ppn=8 #PBS -S /bin/bash #PBS -d ./ #PBS -e ./postprocessing.err #PBS -o ./postprocessing.out### Calculate the number of swarms ### s=`ls -d ./swarm_* | wc -l` swarms=$((s-1))### Create files for Ant-Thony ###for i in $(seq 0 $swarms)doecho "cd swarm_${i}; lgd_generate_conformations.py ../protein.pdb ../dna.pdb gso_100.out 200 > /dev/null 2> /dev/null;" >> generate_lightdock.list;donefor i in $(seq 0 $swarms)doecho "cd swarm_${i}; lgd_cluster_bsas.py gso_100.out > /dev/null 2> /dev/null;" >> cluster_lightdock.list;done### Generate LightDock models ### ant_thony.py -c 8 generate_lightdock.list;### Clustering BSAS (rmsd) within swarm ### ant_thony.py -c 8 cluster_lightdock.list;### Generate ranking files for filtering ### lgd_rank.py $s 100;### Filtering models by >40% of satisfied restraints ### lgd_filter_restraints.py --cutoff 5.0 --fnat 0.4 -lnuc rank_by_scoring.list restraints.list A B#PBS -N 1AZP-post #PBS -q medium #PBS -l nodes=1:ppn=8 #PBS -S /bin/bash #PBS -d ./ #PBS -e ./postprocessing.err #PBS -o ./postprocessing.out### Calculate the number of swarms ### s=`ls -d ./swarm_* | wc -l` swarms=$((s-1))### Create files for Ant-Thony ### for i in $(seq 0 $swarms)doecho "cd swarm_${i}; lgd_generate_conformations.py ../protein.pdb ../dna.pdb gso_100.out 200 > /dev/null 2> /dev/null;" >> generate_lightdock.list;donefor i in $(seq 0 $swarms)doecho "cd swarm_${i}; lgd_cluster_bsas.py gso_100.out > /dev/null 2> /dev/null;" >> cluster_lightdock.list;done### Generate LightDock models ### ant_thony.py -c 8 generate_lightdock.list;### Clustering BSAS (rmsd) within swarm ### ant_thony.py -c 8 cluster_lightdock.list;### Generate ranking files for filtering ### lgd_rank.py $s 100;### Filtering models by >40% of satisfied restraints ### lgd_filter_restraints.py --cutoff 5.0 --fnat 0.4 -lnuc rank_by_scoring.list restraints.list A B -
dz
-
Renumbering a structure 重新编号结构
pdb_reres -1 1ctf.pdb > 1ctf_renumbered.pdb
pdb_reres -1 1ctf.pdb > 1ctf_renumbered.pdbpdb_reres -1 1AZP_A_h.pdb > protein.pdbrm -rf swarm_*
用lightdock 进行Protein-DNA docking prediction。 首先,我们需要确保蛋白质配体的氢原子符合我们在 dna 评分函数中的参数化( dna 评分函数基于 AMBER94 力场)。为此,我们将使用 reduce 软件。请问如何在Linux系统用conda安装 reduce ?我们使用 PDB-Tools(网络服务器或 Python 包)重新编号了蛋白质受体配体的原子:
这里用
