【520特辑】情人节脑影像绘图
祝大家520快乐!
永远爱自己!
1.Brain Net基于节点画爱心

clear all;
clc;t = linspace(0, 2*pi, 30);
x = 16*sin(t).^3;
y = 13*cos(t)-5*cos(2*t)-2*cos(3*t)-cos(4*t);
z = zeros(size(t));[X,Y] = meshgrid(linspace(-10,10,5), linspace(-10,10,5));
X = X(:);
Y = Y(:);
inside = inpolygon(X,Y,x,y);
X = X(inside);
Y = Y(inside);
Z = zeros(size(X));all_x = [x X'];
all_y = [y Y'];
all_z = [z Z'];n_points = length(all_x);
node_data = zeros(n_points, 6);
node_data(:,1:3) = [all_x' all_y' all_z'];
node_data(:,4) = 2;
node_data(:,5) = 2;
node_data(:,6) = 1;dlmwrite('heart_nodes.node', node_data, 'delimiter', '\t', 'precision', 8);adj_matrix = zeros(n_points, n_points);for i = 1:n_pointsfor j = i+1:n_pointsdist = sqrt((all_x(i)-all_x(j))^2 + (all_y(i)-all_y(j))^2);if dist < 6adj_matrix(i,j) = 1;adj_matrix(j,i) = 1;endend
enddlmwrite('heart_edges.edge', adj_matrix, 'delimiter', '\t', 'precision', 8);disp('文件已生成,请在BrainNetViewer中按以下步骤操作:');
disp('1. 打开BrainNetViewer');
disp('2. 选择 Volume/Surface/Edge Files');
disp('3. 在Node File中加载 heart_nodes.node');
disp('4. 在Edge File中加载 heart_edges.edge');
disp('5. 调整Options中的显示参数:');
disp(' - Node Size: 2');
disp(' - Node Color: 选择红色');
disp(' - Edge Color: 选择浅红色');
disp(' - Edge Size: 1');2.创造爱心ROI.nii
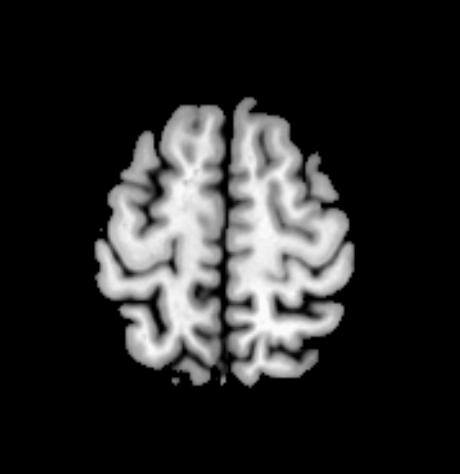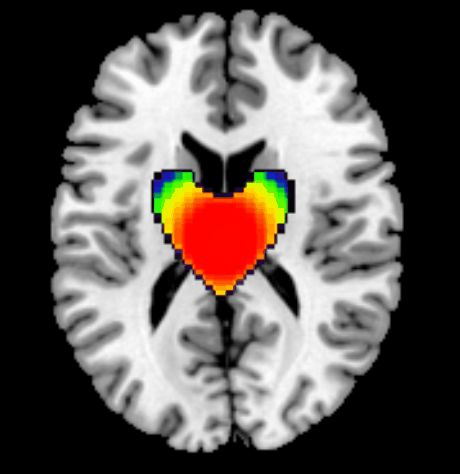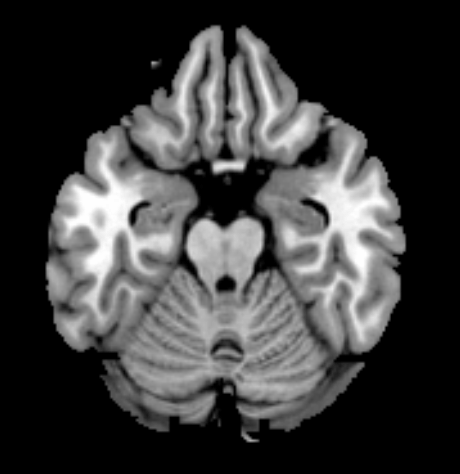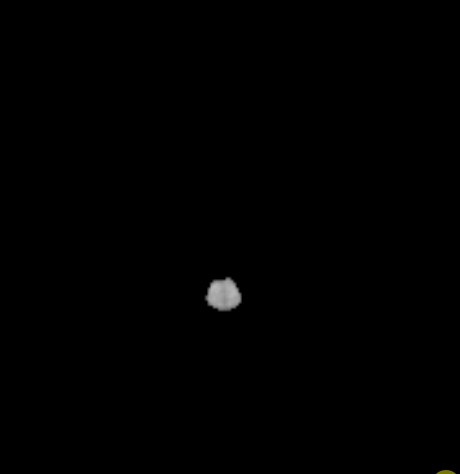
clear all;
clc;dim = [91 109 91];
img = zeros(dim);heart_size = 30;
[x, y, z] = meshgrid(linspace(-1.2,1.2,heart_size), linspace(-1.2,1.2,heart_size), linspace(-1.2,1.2,heart_size));scale = 0.85;
x = scale * x;
y = scale * (y + 0.1);
z = scale * z;heart3d = ((x.^2 + (1.3*y - abs(x)).^2 + z.^2 - 0.85).^3 - x.^2.*z.^3.*0.1 - y.^2.*z.^3.*0.7);
heart_template = heart3d < 0;heart_template = heart_template & (y > -1.0);
top_indent = (x.^2 + (y-1.1).^2 + z.^2) < 0.2;
heart_template = heart_template & ~top_indent;heart_template = permute(heart_template, [2 1 3]);
heart_template = flip(heart_template, 1);
heart_template = flip(heart_template, 2);
heart_template = rot90(heart_template, 1);pos = [46 55 45];half_size = floor(heart_size/2);
x_range = max(1, pos(1)-half_size):min(dim(1), pos(1)+half_size);
y_range = max(1, pos(2)-half_size):min(dim(2), pos(2)+half_size);
z_range = max(1, pos(3)-half_size):min(dim(3), pos(3)+half_size);x_size = length(x_range);
y_size = length(y_range);
z_size = length(z_range);heart_template = heart_template(1:min(heart_size,y_size), 1:min(heart_size,x_size), 1:min(heart_size,z_size));for xi = 1:x_sizefor yi = 1:y_sizefor zi = 1:z_sizeif yi <= size(heart_template,1) && xi <= size(heart_template,2) && zi <= size(heart_template,3)if heart_template(yi,xi,zi)dx = (xi - x_size/2)/(x_size/2);dy = (yi - y_size/2)/(y_size/2);dz = (zi - z_size/2)/(z_size/2);dist = sqrt(dx^2 + dy^2 + dz^2);intensity = max(0, min(1, 1.3 - dist^1.5));current_val = img(x_range(xi), y_range(yi), z_range(zi));img(x_range(xi), y_range(yi), z_range(zi)) = max(current_val, intensity);endendendend
endimg = smooth3(img, 'gaussian', [3 3 3], 0.4);nii = make_nii(img);
nii.hdr.dime.pixdim(2:4) = 2;
nii.hdr.hist.originator(1:3) = [46 64 37];
nii.hdr.hist.qoffset_x = -90;
nii.hdr.hist.qoffset_y = -126;
nii.hdr.hist.qoffset_z = -72;save_nii(nii, 'big_heart_3d_mni_centered_rotated.nii');disp('big_heart_3d_mni_centered_rotated.nii文件已创建完成!');